Please be patient as the page loads
|
PYGB_HUMAN
|
||||||
| SwissProt Accessions | P11216, Q96AK1, Q9NPX8 | Gene names | PYGB | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, brain form (EC 2.4.1.1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PYGB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11216, Q96AK1, Q9NPX8 | Gene names | PYGB | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, brain form (EC 2.4.1.1). | |||||
|
PYGB_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999531 (rank : 2) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CI94, Q8K283 | Gene names | Pygb | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, brain form (EC 2.4.1.1). | |||||
|
PYGL_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.998848 (rank : 3) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06737, O60567, O60752, O60913, Q96G82 | Gene names | PYGL | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, liver form (EC 2.4.1.1). | |||||
|
PYGL_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.998618 (rank : 6) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ET01 | Gene names | Pygl | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, liver form (EC 2.4.1.1). | |||||
|
PYGM_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.998727 (rank : 4) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11217 | Gene names | PYGM | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, muscle form (EC 2.4.1.1) (Myophosphorylase). | |||||
|
PYGM_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.998636 (rank : 5) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WUB3 | Gene names | Pygm | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, muscle form (EC 2.4.1.1) (Myophosphorylase). | |||||
|
SARM1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 7) | NC score | 0.036749 (rank : 8) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
UGGG2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.015735 (rank : 12) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYU1, Q9UFC4 | Gene names | UGCGL2, UGT2 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 2 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase 2) (HUGT2). | |||||
|
PEPD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.041891 (rank : 7) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P12955, Q8TBN9, Q9BT75 | Gene names | PEPD, PRD | |||
|
Domain Architecture |

|
|||||
| Description | Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro dipeptidase) (Proline dipeptidase) (Prolidase) (Imidodipeptidase). | |||||
|
SARM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.030135 (rank : 11) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PDS3, Q5SYG5, Q5SYG6, Q6A054, Q6SZW0, Q8BRI9 | Gene names | Sarm1, Kiaa0524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Tir-1 homolog). | |||||
|
IL7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 11) | NC score | 0.031469 (rank : 10) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13232 | Gene names | IL7 | |||
|
Domain Architecture |
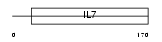
|
|||||
| Description | Interleukin-7 precursor (IL-7). | |||||
|
KIF1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.004349 (rank : 14) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60575, Q9R0B4, Q9WVE5, Q9Z119 | Gene names | Kif1b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B. | |||||
|
BPIL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 13) | NC score | 0.006082 (rank : 13) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C186 | Gene names | Bpil2 | |||
|
Domain Architecture |
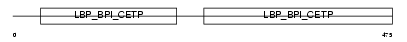
|
|||||
| Description | Bactericidal/permeability-increasing protein-like 2 precursor. | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 14) | NC score | 0.002598 (rank : 15) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
SLC2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 15) | NC score | 0.033613 (rank : 9) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEV8, Q9Y4D6 | Gene names | EXPH5, KIAA0624, SLAC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Slp homolog lacking C2 domains b (Exophilin-5). | |||||
|
PYGB_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11216, Q96AK1, Q9NPX8 | Gene names | PYGB | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, brain form (EC 2.4.1.1). | |||||
|
PYGB_MOUSE
|
||||||
| NC score | 0.999531 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CI94, Q8K283 | Gene names | Pygb | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, brain form (EC 2.4.1.1). | |||||
|
PYGL_HUMAN
|
||||||
| NC score | 0.998848 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06737, O60567, O60752, O60913, Q96G82 | Gene names | PYGL | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, liver form (EC 2.4.1.1). | |||||
|
PYGM_HUMAN
|
||||||
| NC score | 0.998727 (rank : 4) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11217 | Gene names | PYGM | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, muscle form (EC 2.4.1.1) (Myophosphorylase). | |||||
|
PYGM_MOUSE
|
||||||
| NC score | 0.998636 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WUB3 | Gene names | Pygm | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, muscle form (EC 2.4.1.1) (Myophosphorylase). | |||||
|
PYGL_MOUSE
|
||||||
| NC score | 0.998618 (rank : 6) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ET01 | Gene names | Pygl | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, liver form (EC 2.4.1.1). | |||||
|
PEPD_HUMAN
|
||||||
| NC score | 0.041891 (rank : 7) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P12955, Q8TBN9, Q9BT75 | Gene names | PEPD, PRD | |||
|
Domain Architecture |

|
|||||
| Description | Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro dipeptidase) (Proline dipeptidase) (Prolidase) (Imidodipeptidase). | |||||
|
SARM1_HUMAN
|
||||||
| NC score | 0.036749 (rank : 8) | θ value | 0.365318 (rank : 7) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
SLC2B_HUMAN
|
||||||
| NC score | 0.033613 (rank : 9) | θ value | 8.99809 (rank : 15) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEV8, Q9Y4D6 | Gene names | EXPH5, KIAA0624, SLAC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Slp homolog lacking C2 domains b (Exophilin-5). | |||||
|
IL7_HUMAN
|
||||||
| NC score | 0.031469 (rank : 10) | θ value | 6.88961 (rank : 11) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13232 | Gene names | IL7 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-7 precursor (IL-7). | |||||
|
SARM1_MOUSE
|
||||||
| NC score | 0.030135 (rank : 11) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PDS3, Q5SYG5, Q5SYG6, Q6A054, Q6SZW0, Q8BRI9 | Gene names | Sarm1, Kiaa0524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Tir-1 homolog). | |||||
|
UGGG2_HUMAN
|
||||||
| NC score | 0.015735 (rank : 12) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYU1, Q9UFC4 | Gene names | UGCGL2, UGT2 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 2 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase 2) (HUGT2). | |||||
|
BPIL2_MOUSE
|
||||||
| NC score | 0.006082 (rank : 13) | θ value | 8.99809 (rank : 13) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C186 | Gene names | Bpil2 | |||
|
Domain Architecture |

|
|||||
| Description | Bactericidal/permeability-increasing protein-like 2 precursor. | |||||
|
KIF1B_MOUSE
|
||||||
| NC score | 0.004349 (rank : 14) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60575, Q9R0B4, Q9WVE5, Q9Z119 | Gene names | Kif1b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B. | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.002598 (rank : 15) | θ value | 8.99809 (rank : 14) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||