Please be patient as the page loads
|
PLS2_MOUSE
|
||||||
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |
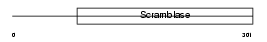
|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PLS2_MOUSE
|
||||||
| θ value | 6.12769e-174 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS1_MOUSE
|
||||||
| θ value | 1.5801e-129 (rank : 2) | NC score | 0.981874 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JJ00, O54730, O54731, Q9D1F8 | Gene names | Plscr1, Tra1b, Tras1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Transplantability-associated protein 1) (TRA1) (NOR1). | |||||
|
PLS1_HUMAN
|
||||||
| θ value | 6.65404e-120 (rank : 3) | NC score | 0.993395 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15162 | Gene names | PLSCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b). | |||||
|
PLS2_HUMAN
|
||||||
| θ value | 6.47494e-99 (rank : 4) | NC score | 0.995449 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS4_HUMAN
|
||||||
| θ value | 1.27248e-70 (rank : 5) | NC score | 0.964534 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
PLS3_MOUSE
|
||||||
| θ value | 2.83479e-70 (rank : 6) | NC score | 0.964434 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS4_MOUSE
|
||||||
| θ value | 1.83746e-69 (rank : 7) | NC score | 0.963925 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
|
PLS3_HUMAN
|
||||||
| θ value | 6.98233e-69 (rank : 8) | NC score | 0.971388 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |
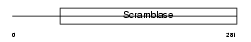
|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
ZN572_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.004609 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3I7, Q8N1Q0 | Gene names | ZNF572 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 572. | |||||
|
ZN334_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.003854 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 778 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCZ1, Q5T6U2, Q9NVW4 | Gene names | ZNF334 | |||
|
Domain Architecture |
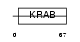
|
|||||
| Description | Zinc finger protein 334. | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.026646 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZN271_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.003178 (rank : 21) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14591, Q96T29, Q9BSX2, Q9UN33, Q9Y5B7 | Gene names | ZNF271, ZNFEB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 271 (Zinc finger protein HZF7) (Zinc finger protein ZNFphex133) (Epstein-Barr virus-induced zinc finger protein) (ZNF-EB) (CT-ZFP48) (Zinc finger protein dp) (ZNF-dp). | |||||
|
KRA47_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.012128 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
CTL4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.009713 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q53GD3, Q5JP84, Q5JQ93, Q658S8, Q8TEW4, Q96C58, Q96K59, Q9Y332 | Gene names | SLC44A4, C6orf29, CTL4, NG22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Choline transporter-like protein 4 (Solute carrier family 44 member 4). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.006314 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
GRAN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.008528 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |
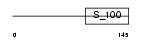
|
|||||
| Description | Grancalcin. | |||||
|
KR410_HUMAN
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.018075 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.034929 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZN605_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.002677 (rank : 22) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86T29, Q86T91 | Gene names | ZNF605 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 605. | |||||
|
KR105_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.005468 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
DAZP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.054031 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.053956 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
PLS2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.12769e-174 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS2_HUMAN
|
||||||
| NC score | 0.995449 (rank : 2) | θ value | 6.47494e-99 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS1_HUMAN
|
||||||
| NC score | 0.993395 (rank : 3) | θ value | 6.65404e-120 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15162 | Gene names | PLSCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b). | |||||
|
PLS1_MOUSE
|
||||||
| NC score | 0.981874 (rank : 4) | θ value | 1.5801e-129 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JJ00, O54730, O54731, Q9D1F8 | Gene names | Plscr1, Tra1b, Tras1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Transplantability-associated protein 1) (TRA1) (NOR1). | |||||
|
PLS3_HUMAN
|
||||||
| NC score | 0.971388 (rank : 5) | θ value | 6.98233e-69 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS4_HUMAN
|
||||||
| NC score | 0.964534 (rank : 6) | θ value | 1.27248e-70 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
PLS3_MOUSE
|
||||||
| NC score | 0.964434 (rank : 7) | θ value | 2.83479e-70 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS4_MOUSE
|
||||||
| NC score | 0.963925 (rank : 8) | θ value | 1.83746e-69 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
|
DAZP2_HUMAN
|
||||||
| NC score | 0.054031 (rank : 9) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| NC score | 0.053956 (rank : 10) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.034929 (rank : 11) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.026646 (rank : 12) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
KR410_HUMAN
|
||||||
| NC score | 0.018075 (rank : 13) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KRA47_HUMAN
|
||||||
| NC score | 0.012128 (rank : 14) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
CTL4_HUMAN
|
||||||
| NC score | 0.009713 (rank : 15) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q53GD3, Q5JP84, Q5JQ93, Q658S8, Q8TEW4, Q96C58, Q96K59, Q9Y332 | Gene names | SLC44A4, C6orf29, CTL4, NG22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Choline transporter-like protein 4 (Solute carrier family 44 member 4). | |||||
|
GRAN_HUMAN
|
||||||
| NC score | 0.008528 (rank : 16) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |

|
|||||
| Description | Grancalcin. | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.006314 (rank : 17) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.005468 (rank : 18) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
ZN572_HUMAN
|
||||||
| NC score | 0.004609 (rank : 19) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3I7, Q8N1Q0 | Gene names | ZNF572 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 572. | |||||
|
ZN334_HUMAN
|
||||||
| NC score | 0.003854 (rank : 20) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 778 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCZ1, Q5T6U2, Q9NVW4 | Gene names | ZNF334 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 334. | |||||
|
ZN271_HUMAN
|
||||||
| NC score | 0.003178 (rank : 21) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14591, Q96T29, Q9BSX2, Q9UN33, Q9Y5B7 | Gene names | ZNF271, ZNFEB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 271 (Zinc finger protein HZF7) (Zinc finger protein ZNFphex133) (Epstein-Barr virus-induced zinc finger protein) (ZNF-EB) (CT-ZFP48) (Zinc finger protein dp) (ZNF-dp). | |||||
|
ZN605_HUMAN
|
||||||
| NC score | 0.002677 (rank : 22) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86T29, Q86T91 | Gene names | ZNF605 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 605. | |||||