Please be patient as the page loads
|
PDCD8_MOUSE
|
||||||
| SwissProt Accessions | Q9Z0X1 | Gene names | Pdcd8, Aif | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 8, mitochondrial precursor (EC 1.-.-.-) (Apoptosis-inducing factor). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PDCD8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998742 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95831, Q6I9X6, Q9Y3I3, Q9Y3I4 | Gene names | PDCD8, AIF | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 8, mitochondrial precursor (EC 1.-.-.-) (Apoptosis-inducing factor). | |||||
|
PDCD8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z0X1 | Gene names | Pdcd8, Aif | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 8, mitochondrial precursor (EC 1.-.-.-) (Apoptosis-inducing factor). | |||||
|
AMID_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 3) | NC score | 0.347785 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BRQ8, Q63Z39 | Gene names | AMID, PRG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-inducing factor-like mitochondrion-associated inducer of death (EC 1.-.-.-) (Apoptosis-inducing factor-homologous mitochondrion-associated inducer of death) (p53-responsive gene 3 protein). | |||||
|
AMID_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 4) | NC score | 0.324857 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUE4, Q8CHZ2 | Gene names | Amid | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-inducing factor-like mitochondrion-associated inducer of death (EC 1.-.-.-) (Apoptosis-inducing factor-homologous mitochondrion-associated inducer of death). | |||||
|
GSHR_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 5) | NC score | 0.102074 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47791, Q7TNC2, Q8BN97, Q8C9Z6 | Gene names | Gsr, Gr1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
GSHR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 6) | NC score | 0.068648 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00390, Q7Z5C9, Q9NP63 | Gene names | GSR, GLUR, GRD1 | |||
|
Domain Architecture |
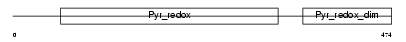
|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 2.36792 (rank : 7) | NC score | 0.015422 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
IRBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 8) | NC score | 0.020228 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49194, Q6B4R6, Q8VD34, Q9R0H8 | Gene names | Rbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interphotoreceptor retinoid-binding protein precursor (IRBP) (Interstitial retinol-binding protein). | |||||
|
ASPM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 9) | NC score | 0.013109 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZT6, Q86UX4, Q8IUL2, Q8IZJ7, Q8IZJ8, Q8IZJ9, Q8N4D1, Q9NVS1, Q9NVT6 | Gene names | ASPM, MCPH5 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein (Abnormal spindle protein homolog) (Asp homolog). | |||||
|
ENOA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 10) | NC score | 0.015002 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06733, P22712, Q16704, Q4TUS4, Q658M5, Q6GMP2, Q71V37, Q7Z3V6, Q8WU71, Q9UM55 | Gene names | ENO1, ENO1L1, MBPB1, MPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-enolase (EC 4.2.1.11) (2-phospho-D-glycerate hydro-lyase) (Non- neural enolase) (NNE) (Enolase 1) (Phosphopyruvate hydratase) (C-myc promoter-binding protein) (MBP-1) (MPB-1) (Plasminogen-binding protein). | |||||
|
K0859_MOUSE
|
||||||
| θ value | 6.88961 (rank : 11) | NC score | 0.027089 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YR5, Q3TFY3, Q69ZX4, Q8BWN2, Q8BX81 | Gene names | Kiaa0859 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0859. | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.017859 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
PDCD8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z0X1 | Gene names | Pdcd8, Aif | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 8, mitochondrial precursor (EC 1.-.-.-) (Apoptosis-inducing factor). | |||||
|
PDCD8_HUMAN
|
||||||
| NC score | 0.998742 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95831, Q6I9X6, Q9Y3I3, Q9Y3I4 | Gene names | PDCD8, AIF | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 8, mitochondrial precursor (EC 1.-.-.-) (Apoptosis-inducing factor). | |||||
|
AMID_HUMAN
|
||||||
| NC score | 0.347785 (rank : 3) | θ value | 5.81887e-07 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BRQ8, Q63Z39 | Gene names | AMID, PRG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-inducing factor-like mitochondrion-associated inducer of death (EC 1.-.-.-) (Apoptosis-inducing factor-homologous mitochondrion-associated inducer of death) (p53-responsive gene 3 protein). | |||||
|
AMID_MOUSE
|
||||||
| NC score | 0.324857 (rank : 4) | θ value | 1.09739e-05 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUE4, Q8CHZ2 | Gene names | Amid | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-inducing factor-like mitochondrion-associated inducer of death (EC 1.-.-.-) (Apoptosis-inducing factor-homologous mitochondrion-associated inducer of death). | |||||
|
GSHR_MOUSE
|
||||||
| NC score | 0.102074 (rank : 5) | θ value | 0.0330416 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47791, Q7TNC2, Q8BN97, Q8C9Z6 | Gene names | Gsr, Gr1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
GSHR_HUMAN
|
||||||
| NC score | 0.068648 (rank : 6) | θ value | 1.81305 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00390, Q7Z5C9, Q9NP63 | Gene names | GSR, GLUR, GRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
K0859_MOUSE
|
||||||
| NC score | 0.027089 (rank : 7) | θ value | 6.88961 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YR5, Q3TFY3, Q69ZX4, Q8BWN2, Q8BX81 | Gene names | Kiaa0859 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0859. | |||||
|
IRBP_MOUSE
|
||||||
| NC score | 0.020228 (rank : 8) | θ value | 5.27518 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49194, Q6B4R6, Q8VD34, Q9R0H8 | Gene names | Rbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interphotoreceptor retinoid-binding protein precursor (IRBP) (Interstitial retinol-binding protein). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.017859 (rank : 9) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.015422 (rank : 10) | θ value | 2.36792 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
ENOA_HUMAN
|
||||||
| NC score | 0.015002 (rank : 11) | θ value | 6.88961 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06733, P22712, Q16704, Q4TUS4, Q658M5, Q6GMP2, Q71V37, Q7Z3V6, Q8WU71, Q9UM55 | Gene names | ENO1, ENO1L1, MBPB1, MPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-enolase (EC 4.2.1.11) (2-phospho-D-glycerate hydro-lyase) (Non- neural enolase) (NNE) (Enolase 1) (Phosphopyruvate hydratase) (C-myc promoter-binding protein) (MBP-1) (MPB-1) (Plasminogen-binding protein). | |||||
|
ASPM_HUMAN
|
||||||
| NC score | 0.013109 (rank : 12) | θ value | 6.88961 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZT6, Q86UX4, Q8IUL2, Q8IZJ7, Q8IZJ8, Q8IZJ9, Q8N4D1, Q9NVS1, Q9NVT6 | Gene names | ASPM, MCPH5 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein (Abnormal spindle protein homolog) (Asp homolog). | |||||