Please be patient as the page loads
|
PA2GA_MOUSE
|
||||||
| SwissProt Accessions | P31482, Q60871 | Gene names | Pla2g2a | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Enhancing factor) (EF). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PA2GA_MOUSE
|
||||||
| θ value | 1.82896e-85 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P31482, Q60871 | Gene names | Pla2g2a | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Enhancing factor) (EF). | |||||
|
PA2GA_HUMAN
|
||||||
| θ value | 1.78386e-56 (rank : 2) | NC score | 0.992260 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P14555, Q6DN24, Q6IBD9, Q9UCD2 | Gene names | PLA2G2A, PLA2B, PLA2L, RASF-A | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Non-pancreatic secretory phospholipase A2) (NPS-PLA2). | |||||
|
PA2GD_HUMAN
|
||||||
| θ value | 1.9192e-34 (rank : 3) | NC score | 0.971524 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UNK4, Q9UK01 | Gene names | PLA2G2D, SPLASH | |||
|
Domain Architecture |

|
|||||
| Description | Group IID secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIID) (GIID sPLA2) (PLA2IID) (sPLA(2)-IID) (Secretory-type PLA, stroma-associated homolog). | |||||
|
PA2GD_MOUSE
|
||||||
| θ value | 1.24399e-33 (rank : 4) | NC score | 0.972506 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVF6, Q3V4B8, Q9JLK0 | Gene names | Pla2g2d, Pla2a2, Splash | |||
|
Domain Architecture |

|
|||||
| Description | Group IID secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIID) (GIID sPLA2) (PLA2IID) (sPLA(2)-IID) (Secretory-type PLA, stroma-associated homolog). | |||||
|
PA2GE_HUMAN
|
||||||
| θ value | 1.79631e-32 (rank : 5) | NC score | 0.970660 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZK7 | Gene names | PLA2G2E | |||
|
Domain Architecture |

|
|||||
| Description | Group IIE secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIE) (GIIE sPLA2) (sPLA(2)-IIE). | |||||
|
PA2GE_MOUSE
|
||||||
| θ value | 4.00176e-32 (rank : 6) | NC score | 0.970414 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QUL3 | Gene names | Pla2g2e | |||
|
Domain Architecture |

|
|||||
| Description | Group IIE secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIE) (GIIE sPLA2) (sPLA(2)-IIE). | |||||
|
PA2G5_HUMAN
|
||||||
| θ value | 7.06379e-29 (rank : 7) | NC score | 0.952946 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P39877, Q8N435 | Gene names | PLA2G5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-dependent phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (PLA2-10) (Group V phospholipase A2). | |||||
|
PA2G5_MOUSE
|
||||||
| θ value | 5.97985e-28 (rank : 8) | NC score | 0.952653 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97391, Q9QZU6 | Gene names | Pla2g5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-dependent phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (PLA2-10) (Group V phospholipase A2). | |||||
|
PA2GX_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 9) | NC score | 0.951113 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15496, Q6NT23 | Gene names | PLA2G10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
|
PA2GX_MOUSE
|
||||||
| θ value | 6.61148e-27 (rank : 10) | NC score | 0.949356 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QXX3, Q9EQK6 | Gene names | Pla2g10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
|
PA2GC_MOUSE
|
||||||
| θ value | 6.18819e-25 (rank : 11) | NC score | 0.956197 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48076 | Gene names | Pla2g2c | |||
|
Domain Architecture |

|
|||||
| Description | Group IIC secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIC) (GIIC sPLA2) (PLA2-8) (14 kDa phospholipase A2). | |||||
|
PA2GF_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 12) | NC score | 0.931835 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZM2, Q5R385, Q8N217, Q9H506 | Gene names | PLA2G2F | |||
|
Domain Architecture |
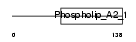
|
|||||
| Description | Group IIF secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIF) (GIIF sPLA2) (sPLA(2)-IIF). | |||||
|
PA2GF_MOUSE
|
||||||
| θ value | 6.40375e-22 (rank : 13) | NC score | 0.930023 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZT4 | Gene names | Pla2g2f | |||
|
Domain Architecture |

|
|||||
| Description | Group IIF secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIF) (GIIF sPLA2) (sPLA(2)-IIF). | |||||
|
PA21B_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 14) | NC score | 0.899233 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0Y2, Q9D7E2, Q9D884 | Gene names | Pla2g1b, Pla2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase) (Group IB phospholipase A2) (PLA2-Ib). | |||||
|
PA21B_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 15) | NC score | 0.896212 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P04054, Q3KPI1 | Gene names | PLA2G1B, PLA2, PLA2A | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase) (Group IB phospholipase A2). | |||||
|
OC90_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 16) | NC score | 0.760648 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02509 | Gene names | OC90, PLA2L | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Phospholipase A2 homolog). | |||||
|
OC90_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 17) | NC score | 0.759984 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0L3, Q9CZ60, Q9Z225 | Gene names | Oc90, Onc-95, Pla2ll | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Otoconin-95) (Oc95). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 18) | NC score | 0.016736 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KR511_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.016927 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.014911 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
CRUM1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.009978 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
KRA58_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.007915 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.013377 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA57_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.009691 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.006772 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
BMR1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.002634 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36894, Q8NEN8 | Gene names | BMPR1A, ACVRLK3, ALK3 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type IA precursor (EC 2.7.11.30) (Serine/threonine-protein kinase receptor R5) (SKR5) (Activin receptor-like kinase 3) (ALK-3) (CD292 antigen). | |||||
|
BMR1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.002655 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36895 | Gene names | Bmpr1a, Acvrlk3, Bmpr | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type IA precursor (EC 2.7.11.30) (Serine/threonine-protein kinase receptor R5) (SKR5) (Activin receptor-like kinase 3) (ALK-3) (BMP-2/BMP-4 receptor). | |||||
|
ADA28_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.003276 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JLN6, Q8K5D2, Q8K5D3 | Gene names | Adam28 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 28 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 28) (Thymic epithelial cell-ADAM) (TECADAM). | |||||
|
PA2GA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.82896e-85 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P31482, Q60871 | Gene names | Pla2g2a | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Enhancing factor) (EF). | |||||
|
PA2GA_HUMAN
|
||||||
| NC score | 0.992260 (rank : 2) | θ value | 1.78386e-56 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P14555, Q6DN24, Q6IBD9, Q9UCD2 | Gene names | PLA2G2A, PLA2B, PLA2L, RASF-A | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Non-pancreatic secretory phospholipase A2) (NPS-PLA2). | |||||
|
PA2GD_MOUSE
|
||||||
| NC score | 0.972506 (rank : 3) | θ value | 1.24399e-33 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVF6, Q3V4B8, Q9JLK0 | Gene names | Pla2g2d, Pla2a2, Splash | |||
|
Domain Architecture |

|
|||||
| Description | Group IID secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIID) (GIID sPLA2) (PLA2IID) (sPLA(2)-IID) (Secretory-type PLA, stroma-associated homolog). | |||||
|
PA2GD_HUMAN
|
||||||
| NC score | 0.971524 (rank : 4) | θ value | 1.9192e-34 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UNK4, Q9UK01 | Gene names | PLA2G2D, SPLASH | |||
|
Domain Architecture |

|
|||||
| Description | Group IID secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIID) (GIID sPLA2) (PLA2IID) (sPLA(2)-IID) (Secretory-type PLA, stroma-associated homolog). | |||||
|
PA2GE_HUMAN
|
||||||
| NC score | 0.970660 (rank : 5) | θ value | 1.79631e-32 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZK7 | Gene names | PLA2G2E | |||
|
Domain Architecture |

|
|||||
| Description | Group IIE secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIE) (GIIE sPLA2) (sPLA(2)-IIE). | |||||
|
PA2GE_MOUSE
|
||||||
| NC score | 0.970414 (rank : 6) | θ value | 4.00176e-32 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QUL3 | Gene names | Pla2g2e | |||
|
Domain Architecture |

|
|||||
| Description | Group IIE secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIE) (GIIE sPLA2) (sPLA(2)-IIE). | |||||
|
PA2GC_MOUSE
|
||||||
| NC score | 0.956197 (rank : 7) | θ value | 6.18819e-25 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48076 | Gene names | Pla2g2c | |||
|
Domain Architecture |

|
|||||
| Description | Group IIC secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIC) (GIIC sPLA2) (PLA2-8) (14 kDa phospholipase A2). | |||||
|
PA2G5_HUMAN
|
||||||
| NC score | 0.952946 (rank : 8) | θ value | 7.06379e-29 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P39877, Q8N435 | Gene names | PLA2G5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-dependent phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (PLA2-10) (Group V phospholipase A2). | |||||
|
PA2G5_MOUSE
|
||||||
| NC score | 0.952653 (rank : 9) | θ value | 5.97985e-28 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97391, Q9QZU6 | Gene names | Pla2g5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-dependent phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (PLA2-10) (Group V phospholipase A2). | |||||
|
PA2GX_HUMAN
|
||||||
| NC score | 0.951113 (rank : 10) | θ value | 3.87602e-27 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15496, Q6NT23 | Gene names | PLA2G10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
|
PA2GX_MOUSE
|
||||||
| NC score | 0.949356 (rank : 11) | θ value | 6.61148e-27 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QXX3, Q9EQK6 | Gene names | Pla2g10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
|
PA2GF_HUMAN
|
||||||
| NC score | 0.931835 (rank : 12) | θ value | 4.01107e-24 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZM2, Q5R385, Q8N217, Q9H506 | Gene names | PLA2G2F | |||
|
Domain Architecture |

|
|||||
| Description | Group IIF secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIF) (GIIF sPLA2) (sPLA(2)-IIF). | |||||
|
PA2GF_MOUSE
|
||||||
| NC score | 0.930023 (rank : 13) | θ value | 6.40375e-22 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZT4 | Gene names | Pla2g2f | |||
|
Domain Architecture |

|
|||||
| Description | Group IIF secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIF) (GIIF sPLA2) (sPLA(2)-IIF). | |||||
|
PA21B_MOUSE
|
||||||
| NC score | 0.899233 (rank : 14) | θ value | 3.88503e-19 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0Y2, Q9D7E2, Q9D884 | Gene names | Pla2g1b, Pla2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase) (Group IB phospholipase A2) (PLA2-Ib). | |||||
|
PA21B_HUMAN
|
||||||
| NC score | 0.896212 (rank : 15) | θ value | 3.28887e-18 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P04054, Q3KPI1 | Gene names | PLA2G1B, PLA2, PLA2A | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase) (Group IB phospholipase A2). | |||||
|
OC90_HUMAN
|
||||||
| NC score | 0.760648 (rank : 16) | θ value | 1.52774e-15 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02509 | Gene names | OC90, PLA2L | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Phospholipase A2 homolog). | |||||
|
OC90_MOUSE
|
||||||
| NC score | 0.759984 (rank : 17) | θ value | 1.68911e-14 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0L3, Q9CZ60, Q9Z225 | Gene names | Oc90, Onc-95, Pla2ll | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Otoconin-95) (Oc95). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.016927 (rank : 18) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.016736 (rank : 19) | θ value | 0.0193708 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.014911 (rank : 20) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.013377 (rank : 21) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
CRUM1_HUMAN
|
||||||
| NC score | 0.009978 (rank : 22) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.009691 (rank : 23) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.007915 (rank : 24) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.006772 (rank : 25) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
ADA28_MOUSE
|
||||||
| NC score | 0.003276 (rank : 26) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JLN6, Q8K5D2, Q8K5D3 | Gene names | Adam28 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 28 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 28) (Thymic epithelial cell-ADAM) (TECADAM). | |||||
|
BMR1A_MOUSE
|
||||||
| NC score | 0.002655 (rank : 27) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36895 | Gene names | Bmpr1a, Acvrlk3, Bmpr | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type IA precursor (EC 2.7.11.30) (Serine/threonine-protein kinase receptor R5) (SKR5) (Activin receptor-like kinase 3) (ALK-3) (BMP-2/BMP-4 receptor). | |||||
|
BMR1A_HUMAN
|
||||||
| NC score | 0.002634 (rank : 28) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36894, Q8NEN8 | Gene names | BMPR1A, ACVRLK3, ALK3 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type IA precursor (EC 2.7.11.30) (Serine/threonine-protein kinase receptor R5) (SKR5) (Activin receptor-like kinase 3) (ALK-3) (CD292 antigen). | |||||