Please be patient as the page loads
|
ODBA_MOUSE
|
||||||
| SwissProt Accessions | P50136 | Gene names | Bckdha | |||
|
Domain Architecture |

|
|||||
| Description | 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial precursor (EC 1.2.4.4) (Branched-chain alpha-keto acid dehydrogenase E1 component alpha chain) (BCKDH E1-alpha). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ODBA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997518 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12694, Q16034, Q16472 | Gene names | BCKDHA | |||
|
Domain Architecture |

|
|||||
| Description | 2-oxoisovalerate dehydrogenase alpha subunit, mitochondrial precursor (EC 1.2.4.4) (Branched-chain alpha-keto acid dehydrogenase E1 component alpha chain) (BCKDH E1-alpha) (BCKDE1A). | |||||
|
ODBA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P50136 | Gene names | Bckdha | |||
|
Domain Architecture |

|
|||||
| Description | 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial precursor (EC 1.2.4.4) (Branched-chain alpha-keto acid dehydrogenase E1 component alpha chain) (BCKDH E1-alpha). | |||||
|
ODPAT_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 3) | NC score | 0.745300 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35487 | Gene names | Pdha2, Pdha-2, Pdhal | |||
|
Domain Architecture |
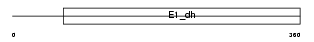
|
|||||
| Description | Pyruvate dehydrogenase E1 component alpha subunit, testis-specific form, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A type II). | |||||
|
ODPA_MOUSE
|
||||||
| θ value | 3.28125e-26 (rank : 4) | NC score | 0.740713 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35486 | Gene names | Pdha1, Pdha-1 | |||
|
Domain Architecture |
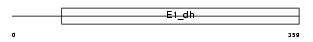
|
|||||
| Description | Pyruvate dehydrogenase E1 component alpha subunit, somatic form, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A type I). | |||||
|
ODPA_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 5) | NC score | 0.735120 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08559, Q9NP12, Q9UBJ8, Q9UBU0, Q9UNG4, Q9UNG5 | Gene names | PDHA1, PHE1A | |||
|
Domain Architecture |
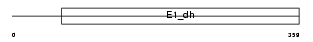
|
|||||
| Description | Pyruvate dehydrogenase E1 component alpha subunit, somatic form, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A type I). | |||||
|
ODPAT_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 6) | NC score | 0.732019 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29803 | Gene names | PDHA2 | |||
|
Domain Architecture |
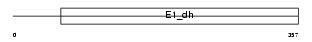
|
|||||
| Description | Pyruvate dehydrogenase E1 component alpha subunit, testis-specific form, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A type II). | |||||
|
TKT_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.138152 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29401, Q8TBA3, Q96HH3 | Gene names | TKT | |||
|
Domain Architecture |

|
|||||
| Description | Transketolase (EC 2.2.1.1) (TK). | |||||
|
TKT_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 8) | NC score | 0.133328 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40142 | Gene names | Tkt | |||
|
Domain Architecture |

|
|||||
| Description | Transketolase (EC 2.2.1.1) (TK) (P68). | |||||
|
EPIPL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.029363 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0W0 | Gene names | Eppk1 | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin. | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.009736 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.027831 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.006834 (rank : 22) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.033626 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 14) | NC score | 0.009462 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.011468 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
NGAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.011021 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJF2, O95174, Q5TFU9 | Gene names | RASAL2, NGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein nGAP (RAS protein activator-like 1). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.024427 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.015576 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.005542 (rank : 23) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.008767 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
SYF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.028317 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D198 | Gene names | Syf2, Cbpin, Gcipip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor SYF2 (CCNDBP1-interactor) (p29) (mp29). | |||||
|
ODO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.050258 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02218 | Gene names | OGDH | |||
|
Domain Architecture |

|
|||||
| Description | 2-oxoglutarate dehydrogenase E1 component, mitochondrial precursor (EC 1.2.4.2) (Alpha-ketoglutarate dehydrogenase). | |||||
|
TKT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.053597 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51854 | Gene names | TKTL1, TKR, TKT2 | |||
|
Domain Architecture |

|
|||||
| Description | Transketolase-like 1 (EC 2.2.1.1) (Transketolase 2) (TK 2) (Transketolase-related protein). | |||||
|
ODBA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P50136 | Gene names | Bckdha | |||
|
Domain Architecture |

|
|||||
| Description | 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial precursor (EC 1.2.4.4) (Branched-chain alpha-keto acid dehydrogenase E1 component alpha chain) (BCKDH E1-alpha). | |||||
|
ODBA_HUMAN
|
||||||
| NC score | 0.997518 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12694, Q16034, Q16472 | Gene names | BCKDHA | |||
|
Domain Architecture |

|
|||||
| Description | 2-oxoisovalerate dehydrogenase alpha subunit, mitochondrial precursor (EC 1.2.4.4) (Branched-chain alpha-keto acid dehydrogenase E1 component alpha chain) (BCKDH E1-alpha) (BCKDE1A). | |||||
|
ODPAT_MOUSE
|
||||||
| NC score | 0.745300 (rank : 3) | θ value | 2.51237e-26 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35487 | Gene names | Pdha2, Pdha-2, Pdhal | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase E1 component alpha subunit, testis-specific form, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A type II). | |||||
|
ODPA_MOUSE
|
||||||
| NC score | 0.740713 (rank : 4) | θ value | 3.28125e-26 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35486 | Gene names | Pdha1, Pdha-1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase E1 component alpha subunit, somatic form, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A type I). | |||||
|
ODPA_HUMAN
|
||||||
| NC score | 0.735120 (rank : 5) | θ value | 1.05554e-24 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08559, Q9NP12, Q9UBJ8, Q9UBU0, Q9UNG4, Q9UNG5 | Gene names | PDHA1, PHE1A | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase E1 component alpha subunit, somatic form, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A type I). | |||||
|
ODPAT_HUMAN
|
||||||
| NC score | 0.732019 (rank : 6) | θ value | 8.93572e-24 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29803 | Gene names | PDHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase E1 component alpha subunit, testis-specific form, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A type II). | |||||
|
TKT_HUMAN
|
||||||
| NC score | 0.138152 (rank : 7) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29401, Q8TBA3, Q96HH3 | Gene names | TKT | |||
|
Domain Architecture |

|
|||||
| Description | Transketolase (EC 2.2.1.1) (TK). | |||||
|
TKT_MOUSE
|
||||||
| NC score | 0.133328 (rank : 8) | θ value | 0.0563607 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40142 | Gene names | Tkt | |||
|
Domain Architecture |

|
|||||
| Description | Transketolase (EC 2.2.1.1) (TK) (P68). | |||||
|
TKT2_HUMAN
|
||||||
| NC score | 0.053597 (rank : 9) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51854 | Gene names | TKTL1, TKR, TKT2 | |||
|
Domain Architecture |

|
|||||
| Description | Transketolase-like 1 (EC 2.2.1.1) (Transketolase 2) (TK 2) (Transketolase-related protein). | |||||
|
ODO1_HUMAN
|
||||||
| NC score | 0.050258 (rank : 10) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02218 | Gene names | OGDH | |||
|
Domain Architecture |

|
|||||
| Description | 2-oxoglutarate dehydrogenase E1 component, mitochondrial precursor (EC 1.2.4.2) (Alpha-ketoglutarate dehydrogenase). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.033626 (rank : 11) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
EPIPL_MOUSE
|
||||||
| NC score | 0.029363 (rank : 12) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0W0 | Gene names | Eppk1 | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin. | |||||
|
SYF2_MOUSE
|
||||||
| NC score | 0.028317 (rank : 13) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D198 | Gene names | Syf2, Cbpin, Gcipip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor SYF2 (CCNDBP1-interactor) (p29) (mp29). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.027831 (rank : 14) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.024427 (rank : 15) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.015576 (rank : 16) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.011468 (rank : 17) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
NGAP_HUMAN
|
||||||
| NC score | 0.011021 (rank : 18) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJF2, O95174, Q5TFU9 | Gene names | RASAL2, NGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein nGAP (RAS protein activator-like 1). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.009736 (rank : 19) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.009462 (rank : 20) | θ value | 5.27518 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.008767 (rank : 21) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.006834 (rank : 22) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.005542 (rank : 23) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||