Please be patient as the page loads
|
NUD13_MOUSE
|
||||||
| SwissProt Accessions | Q8JZU0, Q9CXN4 | Gene names | Nudt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NUD13_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8JZU0, Q9CXN4 | Gene names | Nudt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
|
NUD13_HUMAN
|
||||||
| θ value | 5.21151e-157 (rank : 2) | NC score | 0.996587 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86X67, Q5SQM4, Q5SQM5, Q9Y3X2 | Gene names | NUDT13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
|
NUD12_MOUSE
|
||||||
| θ value | 5.39604e-37 (rank : 3) | NC score | 0.576585 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCN1, Q6PFA5 | Gene names | Nudt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUD12_HUMAN
|
||||||
| θ value | 1.01765e-35 (rank : 4) | NC score | 0.595443 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQG2, Q8TAL7 | Gene names | NUDT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUD15_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 5) | NC score | 0.229294 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BG93 | Gene names | Nudt15, Mth2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15) (MutT homolog 2) (mMTH2). | |||||
|
8ODP_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 6) | NC score | 0.284419 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53368, P97795 | Gene names | Nudt1, Mth1 | |||
|
Domain Architecture |
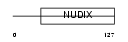
|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
|
8ODP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 7) | NC score | 0.263548 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P36639, Q6P0Y6, Q9UBM0, Q9UBM9 | Gene names | NUDT1, MTH1 | |||
|
Domain Architecture |
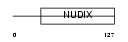
|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
|
K2C8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 8) | NC score | 0.002627 (rank : 12) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
NUDT6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 9) | NC score | 0.031586 (rank : 11) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53370, O95097, Q9UQD9 | Gene names | NUDT6, FGF2AS | |||
|
Domain Architecture |
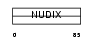
|
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 6 (Nudix motif 6) (Protein GFG) (GFG-1) (Antisense basic fibroblast growth factor). | |||||
|
ANR40_MOUSE
|
||||||
| θ value | θ > 10 (rank : 10) | NC score | 0.052081 (rank : 10) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SUE8, Q8R595, Q9CU71 | Gene names | Ankrd40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 40. | |||||
|
NUD15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 11) | NC score | 0.110210 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NV35, Q6P2C9 | Gene names | NUDT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15). | |||||
|
NUDT5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 12) | NC score | 0.059095 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKX6, Q99KM4 | Gene names | Nudt5 | |||
|
Domain Architecture |
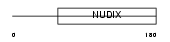
|
|||||
| Description | ADP-sugar pyrophosphatase (EC 3.6.1.13) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 5) (Nudix motif 5). | |||||
|
NUD13_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8JZU0, Q9CXN4 | Gene names | Nudt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
|
NUD13_HUMAN
|
||||||
| NC score | 0.996587 (rank : 2) | θ value | 5.21151e-157 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86X67, Q5SQM4, Q5SQM5, Q9Y3X2 | Gene names | NUDT13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
|
NUD12_HUMAN
|
||||||
| NC score | 0.595443 (rank : 3) | θ value | 1.01765e-35 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQG2, Q8TAL7 | Gene names | NUDT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUD12_MOUSE
|
||||||
| NC score | 0.576585 (rank : 4) | θ value | 5.39604e-37 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCN1, Q6PFA5 | Gene names | Nudt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
8ODP_MOUSE
|
||||||
| NC score | 0.284419 (rank : 5) | θ value | 0.0148317 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53368, P97795 | Gene names | Nudt1, Mth1 | |||
|
Domain Architecture |

|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
|
8ODP_HUMAN
|
||||||
| NC score | 0.263548 (rank : 6) | θ value | 0.125558 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P36639, Q6P0Y6, Q9UBM0, Q9UBM9 | Gene names | NUDT1, MTH1 | |||
|
Domain Architecture |

|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
|
NUD15_MOUSE
|
||||||
| NC score | 0.229294 (rank : 7) | θ value | 0.00509761 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BG93 | Gene names | Nudt15, Mth2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15) (MutT homolog 2) (mMTH2). | |||||
|
NUD15_HUMAN
|
||||||
| NC score | 0.110210 (rank : 8) | θ value | θ > 10 (rank : 11) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NV35, Q6P2C9 | Gene names | NUDT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15). | |||||
|
NUDT5_MOUSE
|
||||||
| NC score | 0.059095 (rank : 9) | θ value | θ > 10 (rank : 12) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKX6, Q99KM4 | Gene names | Nudt5 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-sugar pyrophosphatase (EC 3.6.1.13) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 5) (Nudix motif 5). | |||||
|
ANR40_MOUSE
|
||||||
| NC score | 0.052081 (rank : 10) | θ value | θ > 10 (rank : 10) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SUE8, Q8R595, Q9CU71 | Gene names | Ankrd40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 40. | |||||
|
NUDT6_HUMAN
|
||||||
| NC score | 0.031586 (rank : 11) | θ value | 8.99809 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53370, O95097, Q9UQD9 | Gene names | NUDT6, FGF2AS | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 6 (Nudix motif 6) (Protein GFG) (GFG-1) (Antisense basic fibroblast growth factor). | |||||
|
K2C8_HUMAN
|
||||||
| NC score | 0.002627 (rank : 12) | θ value | 6.88961 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||