Please be patient as the page loads
|
LPH_HUMAN
|
||||||
| SwissProt Accessions | P09848 | Gene names | LCT, LPH | |||
|
Domain Architecture |

|
|||||
| Description | Lactase-phlorizin hydrolase precursor (Lactase-glycosylceramidase) [Includes: Lactase (EC 3.2.1.108); Phlorizin hydrolase (EC 3.2.1.62)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KLOTB_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.964339 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99N32, Q920J2, Q99N31 | Gene names | Klb, Betakl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta klotho (BetaKlotho) (Klotho beta-like protein). | |||||
|
KLOT_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.965022 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UEF7, Q5VZ95, Q96KV5, Q96KW5, Q9UEI9, Q9Y4F0 | Gene names | KL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Klotho precursor (EC 3.2.1.31) [Contains: Klotho peptide]. | |||||
|
KLOT_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.965089 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35082, O70175, O70621 | Gene names | Kl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Klotho precursor (EC 3.2.1.31) [Contains: Klotho peptide]. | |||||
|
LPH_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09848 | Gene names | LCT, LPH | |||
|
Domain Architecture |

|
|||||
| Description | Lactase-phlorizin hydrolase precursor (Lactase-glycosylceramidase) [Includes: Lactase (EC 3.2.1.108); Phlorizin hydrolase (EC 3.2.1.62)]. | |||||
|
KLOTB_HUMAN
|
||||||
| θ value | 3.24166e-183 (rank : 5) | NC score | 0.963502 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86Z14 | Gene names | KLB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta klotho (BetaKlotho) (Klotho beta-like protein). | |||||
|
LCTL_HUMAN
|
||||||
| θ value | 1.74296e-136 (rank : 6) | NC score | 0.961316 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UWM7 | Gene names | LCTL, KLPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lactase-like protein precursor (Klotho/lactase-phlorizin hydrolase- related protein). | |||||
|
LCTL_MOUSE
|
||||||
| θ value | 1.52691e-132 (rank : 7) | NC score | 0.960013 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K1F9, Q8BPR1, Q8K2M9 | Gene names | Lctl, Klph | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lactase-like protein precursor (Klotho/lactase-phlorizin hydrolase- related protein). | |||||
|
GBA3_HUMAN
|
||||||
| θ value | 8.67031e-128 (rank : 8) | NC score | 0.969467 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H227, Q3MIH4, Q53GG8, Q6NSF4, Q8NHT8, Q9H3T4, Q9H4C6 | Gene names | GBA3, CBG, CBGL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic beta-glucosidase (EC 3.2.1.21) (Cytosolic beta-glucosidase- like protein 1). | |||||
|
ANKH_HUMAN
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.046690 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCJ1, Q9NQW2 | Gene names | ANKH, KIAA1581 | |||
|
Domain Architecture |
|
|||||
| Description | Progressive ankylosis protein homolog (ANK). | |||||
|
DYHC_HUMAN
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.022464 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
ANKH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.044284 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHZ2, O35138, O35139 | Gene names | Ankh, Ank | |||
|
Domain Architecture |
|
|||||
| Description | Progressive ankylosis protein (Fn54 protein). | |||||
|
DYHC_MOUSE
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.021575 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
NKRF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.014556 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
STON1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.012387 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
TFAM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.014311 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
ADNP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.011697 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H2P0, O94881, Q9UG34 | Gene names | ADNP, KIAA0784 | |||
|
Domain Architecture |

|
|||||
| Description | Activity-dependent neuroprotector (Activity-dependent neuroprotective protein). | |||||
|
CPNE5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.011831 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCH3 | Gene names | CPNE5, KIAA1599 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-5 (Copine V). | |||||
|
CPNE5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.011816 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8JZW4 | Gene names | Cpne5 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-5 (Copine V). | |||||
|
HPLN4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.006773 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80WM4, Q80XX2 | Gene names | Hapln4, Bral2, Lpr4 | |||
|
Domain Architecture |
|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2) (Link protein 4). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.007469 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
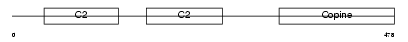
CPNE8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.011230 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DC53, Q9CUZ8 | Gene names | Cpne8 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
DCNL4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.010689 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CCA0 | Gene names | Dcun1d4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCN1-like protein 4 (Defective in cullin neddylation protein 1-like protein 4) (DCUN1 domain-containing protein 4). | |||||
|
FXL13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.008319 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEE6, Q6UVW7, Q6UVW8, Q75MN5, Q86UJ5, Q8N7Y4, Q8TCL2, Q8WUF9, Q8WUG0 | Gene names | FBXL13, FBL13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 13 (F-box and leucine-rich repeat protein 13). | |||||
|
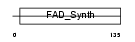
RIFK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.015703 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969G6, Q9NUT7 | Gene names | RFK | |||
|
Domain Architecture |
|
|||||
| Description | Riboflavin kinase (EC 2.7.1.26) (ATP:riboflavin 5'-phosphotransferase) (Flavokinase). | |||||
|
SGCD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.008875 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P82347 | Gene names | Sgcd | |||
|
Domain Architecture |

|
|||||
| Description | Delta-sarcoglycan (SG-delta) (35 kDa dystrophin-associated glycoprotein) (35DAG). | |||||
|
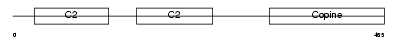
CPNE8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.010643 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86YQ8, Q2TB41 | Gene names | CPNE8 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
MEOX2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.001396 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32443, Q544T6 | Gene names | Meox2, Gax, Mox-2, Mox2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-2 (Mesenchyme homeobox 2). | |||||
|
LPH_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09848 | Gene names | LCT, LPH | |||
|
Domain Architecture |

|
|||||
| Description | Lactase-phlorizin hydrolase precursor (Lactase-glycosylceramidase) [Includes: Lactase (EC 3.2.1.108); Phlorizin hydrolase (EC 3.2.1.62)]. | |||||
|
GBA3_HUMAN
|
||||||
| NC score | 0.969467 (rank : 2) | θ value | 8.67031e-128 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H227, Q3MIH4, Q53GG8, Q6NSF4, Q8NHT8, Q9H3T4, Q9H4C6 | Gene names | GBA3, CBG, CBGL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic beta-glucosidase (EC 3.2.1.21) (Cytosolic beta-glucosidase- like protein 1). | |||||
|
KLOT_MOUSE
|
||||||
| NC score | 0.965089 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35082, O70175, O70621 | Gene names | Kl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Klotho precursor (EC 3.2.1.31) [Contains: Klotho peptide]. | |||||
|
KLOT_HUMAN
|
||||||
| NC score | 0.965022 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UEF7, Q5VZ95, Q96KV5, Q96KW5, Q9UEI9, Q9Y4F0 | Gene names | KL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Klotho precursor (EC 3.2.1.31) [Contains: Klotho peptide]. | |||||
|
KLOTB_MOUSE
|
||||||
| NC score | 0.964339 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99N32, Q920J2, Q99N31 | Gene names | Klb, Betakl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta klotho (BetaKlotho) (Klotho beta-like protein). | |||||
|
KLOTB_HUMAN
|
||||||
| NC score | 0.963502 (rank : 6) | θ value | 3.24166e-183 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86Z14 | Gene names | KLB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta klotho (BetaKlotho) (Klotho beta-like protein). | |||||
|
LCTL_HUMAN
|
||||||
| NC score | 0.961316 (rank : 7) | θ value | 1.74296e-136 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UWM7 | Gene names | LCTL, KLPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lactase-like protein precursor (Klotho/lactase-phlorizin hydrolase- related protein). | |||||
|
LCTL_MOUSE
|
||||||
| NC score | 0.960013 (rank : 8) | θ value | 1.52691e-132 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K1F9, Q8BPR1, Q8K2M9 | Gene names | Lctl, Klph | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lactase-like protein precursor (Klotho/lactase-phlorizin hydrolase- related protein). | |||||
|
ANKH_HUMAN
|
||||||
| NC score | 0.046690 (rank : 9) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCJ1, Q9NQW2 | Gene names | ANKH, KIAA1581 | |||
|
Domain Architecture |

|
|||||
| Description | Progressive ankylosis protein homolog (ANK). | |||||
|
ANKH_MOUSE
|
||||||
| NC score | 0.044284 (rank : 10) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHZ2, O35138, O35139 | Gene names | Ankh, Ank | |||
|
Domain Architecture |

|
|||||
| Description | Progressive ankylosis protein (Fn54 protein). | |||||
|
DYHC_HUMAN
|
||||||
| NC score | 0.022464 (rank : 11) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_MOUSE
|
||||||
| NC score | 0.021575 (rank : 12) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
RIFK_HUMAN
|
||||||
| NC score | 0.015703 (rank : 13) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969G6, Q9NUT7 | Gene names | RFK | |||
|
Domain Architecture |

|
|||||
| Description | Riboflavin kinase (EC 2.7.1.26) (ATP:riboflavin 5'-phosphotransferase) (Flavokinase). | |||||
|
NKRF_HUMAN
|
||||||
| NC score | 0.014556 (rank : 14) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
TFAM_HUMAN
|
||||||
| NC score | 0.014311 (rank : 15) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
STON1_MOUSE
|
||||||
| NC score | 0.012387 (rank : 16) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
CPNE5_HUMAN
|
||||||
| NC score | 0.011831 (rank : 17) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCH3 | Gene names | CPNE5, KIAA1599 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-5 (Copine V). | |||||
|
CPNE5_MOUSE
|
||||||
| NC score | 0.011816 (rank : 18) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8JZW4 | Gene names | Cpne5 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-5 (Copine V). | |||||
|
ADNP_HUMAN
|
||||||
| NC score | 0.011697 (rank : 19) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H2P0, O94881, Q9UG34 | Gene names | ADNP, KIAA0784 | |||
|
Domain Architecture |

|
|||||
| Description | Activity-dependent neuroprotector (Activity-dependent neuroprotective protein). | |||||
|
CPNE8_MOUSE
|
||||||
| NC score | 0.011230 (rank : 20) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DC53, Q9CUZ8 | Gene names | Cpne8 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
DCNL4_MOUSE
|
||||||
| NC score | 0.010689 (rank : 21) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CCA0 | Gene names | Dcun1d4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCN1-like protein 4 (Defective in cullin neddylation protein 1-like protein 4) (DCUN1 domain-containing protein 4). | |||||
|
CPNE8_HUMAN
|
||||||
| NC score | 0.010643 (rank : 22) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86YQ8, Q2TB41 | Gene names | CPNE8 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
SGCD_MOUSE
|
||||||
| NC score | 0.008875 (rank : 23) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P82347 | Gene names | Sgcd | |||
|
Domain Architecture |

|
|||||
| Description | Delta-sarcoglycan (SG-delta) (35 kDa dystrophin-associated glycoprotein) (35DAG). | |||||
|
FXL13_HUMAN
|
||||||
| NC score | 0.008319 (rank : 24) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEE6, Q6UVW7, Q6UVW8, Q75MN5, Q86UJ5, Q8N7Y4, Q8TCL2, Q8WUF9, Q8WUG0 | Gene names | FBXL13, FBL13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 13 (F-box and leucine-rich repeat protein 13). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.007469 (rank : 25) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
HPLN4_MOUSE
|
||||||
| NC score | 0.006773 (rank : 26) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80WM4, Q80XX2 | Gene names | Hapln4, Bral2, Lpr4 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2) (Link protein 4). | |||||
|
MEOX2_MOUSE
|
||||||
| NC score | 0.001396 (rank : 27) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32443, Q544T6 | Gene names | Meox2, Gax, Mox-2, Mox2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-2 (Mesenchyme homeobox 2). | |||||