Please be patient as the page loads
|
KCMB2_HUMAN
|
||||||
| SwissProt Accessions | Q9Y691 | Gene names | KCNMB2 | |||
|
Domain Architecture |
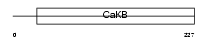
|
|||||
| Description | Calcium-activated potassium channel subunit beta 2 (Calcium-activated potassium channel, subfamily M subunit beta 2) (Maxi K channel subunit beta 2) (BK channel subunit beta 2) (Slo-beta 2) (K(VCA)beta 2) (Charybdotoxin receptor subunit beta 2) (BKbeta2) (Hbeta2) (Hbeta3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCMB2_HUMAN
|
||||||
| θ value | 2.27638e-136 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y691 | Gene names | KCNMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 2 (Calcium-activated potassium channel, subfamily M subunit beta 2) (Maxi K channel subunit beta 2) (BK channel subunit beta 2) (Slo-beta 2) (K(VCA)beta 2) (Charybdotoxin receptor subunit beta 2) (BKbeta2) (Hbeta2) (Hbeta3). | |||||
|
KCMB2_MOUSE
|
||||||
| θ value | 1.09426e-130 (rank : 2) | NC score | 0.998787 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CZM9 | Gene names | Kcnmb2 | |||
|
Domain Architecture |
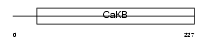
|
|||||
| Description | Calcium-activated potassium channel subunit beta 2 (Calcium-activated potassium channel, subfamily M subunit beta 2) (Maxi K channel subunit beta 2) (BK channel subunit beta 2) (Slo-beta 2) (K(VCA)beta 2) (Charybdotoxin receptor subunit beta 2) (BKbeta2). | |||||
|
KCMB3_HUMAN
|
||||||
| θ value | 1.28137e-46 (rank : 3) | NC score | 0.934609 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPA1, Q9NPG7, Q9NRM9, Q9UHN3 | Gene names | KCNMB3 | |||
|
Domain Architecture |
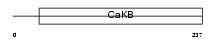
|
|||||
| Description | Calcium-activated potassium channel subunit beta 3 (Calcium-activated potassium channel, subfamily M subunit beta 3) (Maxi K channel subunit beta 3) (BK channel subunit beta 3) (Slo-beta 3) (K(VCA)beta 3) (Charybdotoxin receptor subunit beta 3) (BKbeta3) (Hbeta3). | |||||
|
KCMB1_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 4) | NC score | 0.904284 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16558, O00707, O00708, P78475, Q8TAX3, Q93005 | Gene names | KCNMB1 | |||
|
Domain Architecture |
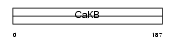
|
|||||
| Description | Calcium-activated potassium channel subunit beta 1 (Calcium-activated potassium channel, subfamily M subunit beta 1) (Maxi K channel subunit beta 1) (BK channel subunit beta 1) (Slo-beta 1) (K(VCA)beta 1) (Charybdotoxin receptor subunit beta 1) (BKbeta1) (Hbeta1) (Calcium- activated potassium channel subunit beta) (BKbeta) (Slo-beta). | |||||
|
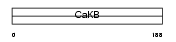
KCMB1_MOUSE
|
||||||
| θ value | 1.16164e-39 (rank : 5) | NC score | 0.901029 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CAE3, O35336, O35645 | Gene names | Kcnmb1 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-activated potassium channel subunit beta 1 (Calcium-activated potassium channel, subfamily M subunit beta 1) (Maxi K channel subunit beta 1) (BK channel subunit beta 1) (Slo-beta 1) (K(VCA)beta 1) (Charybdotoxin receptor subunit beta 1) (BKbeta1) (Calcium-activated potassium channel subunit beta) (BKbeta) (Slo-beta). | |||||
|
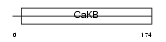
KCMB4_HUMAN
|
||||||
| θ value | 7.80994e-28 (rank : 6) | NC score | 0.826981 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86W47, Q8IVR3, Q9NPA4, Q9P0G5 | Gene names | KCNMB4 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-activated potassium channel subunit beta 4 (Calcium-activated potassium channel, subfamily M subunit beta 4) (Maxi K channel subunit beta 4) (BK channel subunit beta 4) (Slo-beta 4) (K(VCA)beta 4) (Charybdotoxin receptor subunit beta 4) (BKbeta4) (Hbeta4). | |||||
|
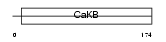
KCMB4_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 7) | NC score | 0.820827 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JIN6 | Gene names | Kcnmb4 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-activated potassium channel subunit beta 4 (Calcium-activated potassium channel, subfamily M subunit beta 4) (Maxi K channel subunit beta 4) (BK channel subunit beta 4) (Slo-beta 4) (K(VCA)beta 4) (Charybdotoxin receptor subunit beta 4) (BKbeta4). | |||||
|
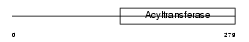
GNPAT_MOUSE
|
||||||
| θ value | 1.38821 (rank : 8) | NC score | 0.029955 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98192, Q9WUT6 | Gene names | Gnpat, Dhapat | |||
|
Domain Architecture |
|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
|
ZN689_HUMAN
|
||||||
| θ value | 1.81305 (rank : 9) | NC score | 0.002458 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96CS4, Q658J5 | Gene names | ZNF689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 689. | |||||
|
ZN689_MOUSE
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.002218 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BKK5, Q8C1C6 | Gene names | Znf689, Zfp689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 689. | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.009236 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
SFXN4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.014352 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P4A7, Q6WSU4, Q86TD9 | Gene names | SFXN4, BCRM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sideroflexin-4 (Breast cancer resistance marker 1). | |||||
|
ZN572_HUMAN
|
||||||
| θ value | 8.99809 (rank : 13) | NC score | 0.000761 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z3I7, Q8N1Q0 | Gene names | ZNF572 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 572. | |||||
|
KCMB2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.27638e-136 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y691 | Gene names | KCNMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 2 (Calcium-activated potassium channel, subfamily M subunit beta 2) (Maxi K channel subunit beta 2) (BK channel subunit beta 2) (Slo-beta 2) (K(VCA)beta 2) (Charybdotoxin receptor subunit beta 2) (BKbeta2) (Hbeta2) (Hbeta3). | |||||
|
KCMB2_MOUSE
|
||||||
| NC score | 0.998787 (rank : 2) | θ value | 1.09426e-130 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CZM9 | Gene names | Kcnmb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 2 (Calcium-activated potassium channel, subfamily M subunit beta 2) (Maxi K channel subunit beta 2) (BK channel subunit beta 2) (Slo-beta 2) (K(VCA)beta 2) (Charybdotoxin receptor subunit beta 2) (BKbeta2). | |||||
|
KCMB3_HUMAN
|
||||||
| NC score | 0.934609 (rank : 3) | θ value | 1.28137e-46 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPA1, Q9NPG7, Q9NRM9, Q9UHN3 | Gene names | KCNMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 3 (Calcium-activated potassium channel, subfamily M subunit beta 3) (Maxi K channel subunit beta 3) (BK channel subunit beta 3) (Slo-beta 3) (K(VCA)beta 3) (Charybdotoxin receptor subunit beta 3) (BKbeta3) (Hbeta3). | |||||
|
KCMB1_HUMAN
|
||||||
| NC score | 0.904284 (rank : 4) | θ value | 2.34062e-40 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16558, O00707, O00708, P78475, Q8TAX3, Q93005 | Gene names | KCNMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 1 (Calcium-activated potassium channel, subfamily M subunit beta 1) (Maxi K channel subunit beta 1) (BK channel subunit beta 1) (Slo-beta 1) (K(VCA)beta 1) (Charybdotoxin receptor subunit beta 1) (BKbeta1) (Hbeta1) (Calcium- activated potassium channel subunit beta) (BKbeta) (Slo-beta). | |||||
|
KCMB1_MOUSE
|
||||||
| NC score | 0.901029 (rank : 5) | θ value | 1.16164e-39 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CAE3, O35336, O35645 | Gene names | Kcnmb1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 1 (Calcium-activated potassium channel, subfamily M subunit beta 1) (Maxi K channel subunit beta 1) (BK channel subunit beta 1) (Slo-beta 1) (K(VCA)beta 1) (Charybdotoxin receptor subunit beta 1) (BKbeta1) (Calcium-activated potassium channel subunit beta) (BKbeta) (Slo-beta). | |||||
|
KCMB4_HUMAN
|
||||||
| NC score | 0.826981 (rank : 6) | θ value | 7.80994e-28 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86W47, Q8IVR3, Q9NPA4, Q9P0G5 | Gene names | KCNMB4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 4 (Calcium-activated potassium channel, subfamily M subunit beta 4) (Maxi K channel subunit beta 4) (BK channel subunit beta 4) (Slo-beta 4) (K(VCA)beta 4) (Charybdotoxin receptor subunit beta 4) (BKbeta4) (Hbeta4). | |||||
|
KCMB4_MOUSE
|
||||||
| NC score | 0.820827 (rank : 7) | θ value | 8.63488e-27 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JIN6 | Gene names | Kcnmb4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 4 (Calcium-activated potassium channel, subfamily M subunit beta 4) (Maxi K channel subunit beta 4) (BK channel subunit beta 4) (Slo-beta 4) (K(VCA)beta 4) (Charybdotoxin receptor subunit beta 4) (BKbeta4). | |||||
|
GNPAT_MOUSE
|
||||||
| NC score | 0.029955 (rank : 8) | θ value | 1.38821 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98192, Q9WUT6 | Gene names | Gnpat, Dhapat | |||
|
Domain Architecture |

|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
|
SFXN4_HUMAN
|
||||||
| NC score | 0.014352 (rank : 9) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P4A7, Q6WSU4, Q86TD9 | Gene names | SFXN4, BCRM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sideroflexin-4 (Breast cancer resistance marker 1). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.009236 (rank : 10) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZN689_HUMAN
|
||||||
| NC score | 0.002458 (rank : 11) | θ value | 1.81305 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96CS4, Q658J5 | Gene names | ZNF689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 689. | |||||
|
ZN689_MOUSE
|
||||||
| NC score | 0.002218 (rank : 12) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BKK5, Q8C1C6 | Gene names | Znf689, Zfp689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 689. | |||||
|
ZN572_HUMAN
|
||||||
| NC score | 0.000761 (rank : 13) | θ value | 8.99809 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z3I7, Q8N1Q0 | Gene names | ZNF572 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 572. | |||||