Please be patient as the page loads
|
IRK8_HUMAN
|
||||||
| SwissProt Accessions | Q15842, O00657 | Gene names | KCNJ8 | |||
|
Domain Architecture |
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 8 (Potassium channel, inwardly rectifying subfamily J member 8) (Inwardly rectifier K(+) channel Kir6.1) (uKATP-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IRK8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15842, O00657 | Gene names | KCNJ8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 8 (Potassium channel, inwardly rectifying subfamily J member 8) (Inwardly rectifier K(+) channel Kir6.1) (uKATP-1). | |||||
|
IRK8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999852 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97794 | Gene names | Kcnj8 | |||
|
Domain Architecture |
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 8 (Potassium channel, inwardly rectifying subfamily J member 8) (Inwardly rectifier K(+) channel Kir6.1) (uKATP-1). | |||||
|
IRK11_HUMAN
|
||||||
| θ value | 2.19465e-147 (rank : 3) | NC score | 0.998328 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14654, Q58EX3 | Gene names | KCNJ11 | |||
|
Domain Architecture |
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 11 (Potassium channel, inwardly rectifying subfamily J member 11) (Inward rectifier K(+) channel Kir6.2) (IKATP). | |||||
|
IRK11_MOUSE
|
||||||
| θ value | 4.13891e-146 (rank : 4) | NC score | 0.998286 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61743, Q9QX21 | Gene names | Kcnj11 | |||
|
Domain Architecture |
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 11 (Potassium channel, inwardly rectifying subfamily J member 11) (Inward rectifier K(+) channel Kir6.2). | |||||
|
IRK6_HUMAN
|
||||||
| θ value | 5.1304e-96 (rank : 5) | NC score | 0.989366 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48051, Q53WW6 | Gene names | KCNJ6, GIRK2, KATP2, KCNJ7 | |||
|
Domain Architecture |
|
|||||
| Description | G protein-activated inward rectifier potassium channel 2 (GIRK2) (Potassium channel, inwardly rectifying subfamily J member 6) (Inward rectifier K(+) channel Kir3.2) (KATP-2) (BIR1). | |||||
|
IRK5_HUMAN
|
||||||
| θ value | 2.63489e-92 (rank : 6) | NC score | 0.988765 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48544, Q6DK13, Q6DK14, Q92807 | Gene names | KCNJ5, GIRK4 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 4 (GIRK4) (Potassium channel, inwardly rectifying subfamily J member 5) (Inward rectifier K(+) channel Kir3.4) (Heart KATP channel) (KATP-1) (Cardiac inward rectifier) (CIR). | |||||
|
IRK2_HUMAN
|
||||||
| θ value | 1.00126e-91 (rank : 7) | NC score | 0.986120 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P63252, O15110, P48049 | Gene names | KCNJ2, HIRK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inward rectifier potassium channel 2 (Potassium channel, inwardly rectifying subfamily J member 2) (Inward rectifier K(+) channel Kir2.1) (Cardiac inward rectifier potassium channel) (IRK1). | |||||
|
IRK2_MOUSE
|
||||||
| θ value | 2.91321e-91 (rank : 8) | NC score | 0.985806 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35561 | Gene names | Kcnj2, Irk1 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 2 (Potassium channel, inwardly rectifying subfamily J member 2) (Inward rectifier K(+) channel Kir2.1). | |||||
|
IRK9_MOUSE
|
||||||
| θ value | 8.47616e-91 (rank : 9) | NC score | 0.990243 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48543, Q9WUE1 | Gene names | Kcnj9, Girk3 | |||
|
Domain Architecture |
|
|||||
| Description | G protein-activated inward rectifier potassium channel 3 (GIRK3) (Potassium channel, inwardly rectifying subfamily J member 9) (Inwardly rectifier K(+) channel Kir3.3). | |||||
|
IRK5_MOUSE
|
||||||
| θ value | 1.88829e-90 (rank : 10) | NC score | 0.988777 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48545, P97508 | Gene names | Kcnj5, Girk4 | |||
|
Domain Architecture |
|
|||||
| Description | G protein-activated inward rectifier potassium channel 4 (GIRK4) (Potassium channel, inwardly rectifying subfamily J member 5) (Inward rectifier K(+) channel Kir3.4) (Heart KATP channel) (KATP-1) (Cardiac inward rectifier) (CIR). | |||||
|
IRK3_MOUSE
|
||||||
| θ value | 3.22093e-90 (rank : 11) | NC score | 0.988225 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P63250, P35562 | Gene names | Kcnj3, Girk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-activated inward rectifier potassium channel 1 (GIRK1) (Potassium channel, inwardly rectifying subfamily J member 3) (Inward rectifier K(+) channel Kir3.1). | |||||
|
IRK9_HUMAN
|
||||||
| θ value | 3.22093e-90 (rank : 12) | NC score | 0.990218 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92806, Q5JW75 | Gene names | KCNJ9, GIRK3 | |||
|
Domain Architecture |
|
|||||
| Description | G protein-activated inward rectifier potassium channel 3 (GIRK3) (Potassium channel, inwardly rectifying subfamily J member 9) (Inwardly rectifier K(+) channel Kir3.3). | |||||
|
IRK3_HUMAN
|
||||||
| θ value | 4.20666e-90 (rank : 13) | NC score | 0.988195 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48549, Q8TBI0 | Gene names | KCNJ3, GIRK1 | |||
|
Domain Architecture |
|
|||||
| Description | G protein-activated inward rectifier potassium channel 1 (GIRK1) (Potassium channel, inwardly rectifying subfamily J member 3) (Inward rectifier K(+) channel Kir3.1). | |||||
|
IRK6_MOUSE
|
||||||
| θ value | 6.07444e-89 (rank : 14) | NC score | 0.988865 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48542, O70290, P70216, P70306, P70307, P70308, P70309, P70454, Q9QYH5 | Gene names | Kcnj6, Girk2, Kcnj7, W | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 2 (GIRK2) (Potassium channel, inwardly rectifying subfamily J member 6) (Inward rectifier K(+) channel Kir3.2). | |||||
|
IRK4_HUMAN
|
||||||
| θ value | 9.69798e-87 (rank : 15) | NC score | 0.987986 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48050 | Gene names | KCNJ4 | |||
|
Domain Architecture |
|
|||||
| Description | Inward rectifier potassium channel 4 (Potassium channel, inwardly rectifying subfamily J member 4) (Inward rectifier K(+) channel Kir2.3) (Hippocampal inward rectifier) (HIR) (HRK1) (HIRK2). | |||||
|
IRK4_MOUSE
|
||||||
| θ value | 2.82168e-86 (rank : 16) | NC score | 0.987933 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52189 | Gene names | Kcnj4, Irk3 | |||
|
Domain Architecture |
|
|||||
| Description | Inward rectifier potassium channel 4 (Potassium channel, inwardly rectifying subfamily J member 4) (Inward rectifier K(+) channel Kir2.3) (IRK3). | |||||
|
IRK12_HUMAN
|
||||||
| θ value | 1.54831e-84 (rank : 17) | NC score | 0.985703 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14500, O43401, Q8NG63 | Gene names | KCNJ12, IRK2 | |||
|
Domain Architecture |
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 12 (Potassium channel, inwardly rectifying subfamily J member 12) (Inward rectifier K(+) channel Kir2.2) (IRK2). | |||||
|
IRK12_MOUSE
|
||||||
| θ value | 5.88354e-84 (rank : 18) | NC score | 0.985515 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52187, Q9QYF2 | Gene names | Kcnj12, Irk2 | |||
|
Domain Architecture |
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 12 (Potassium channel, inwardly rectifying subfamily J member 12) (Inward rectifier K(+) channel Kir2.2) (IRK2). | |||||
|
IRKI_HUMAN
|
||||||
| θ value | 1.35638e-80 (rank : 19) | NC score | 0.985312 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15756 | Gene names | KCNJN1 | |||
|
Domain Architecture |
|
|||||
| Description | Inward rectifying K(+) channel negative regulator Kir2.2v. | |||||
|
IRK14_HUMAN
|
||||||
| θ value | 2.02684e-76 (rank : 20) | NC score | 0.985697 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UNX9 | Gene names | KCNJ14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-sensitive inward rectifier potassium channel 14 (Potassium channel, inwardly rectifying subfamily J member 14) (Inward rectifier K(+) channel Kir2.4) (IRK4). | |||||
|
IRK14_MOUSE
|
||||||
| θ value | 5.8972e-76 (rank : 21) | NC score | 0.985538 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8JZN3, Q8BMK3, Q8BXM0 | Gene names | Kcnj14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-sensitive inward rectifier potassium channel 14 (Potassium channel, inwardly rectifying subfamily J member 14) (Inward rectifier K(+) channel Kir2.4) (IRK4). | |||||
|
IRK1_MOUSE
|
||||||
| θ value | 4.99229e-75 (rank : 22) | NC score | 0.985627 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88335 | Gene names | Kcnj1 | |||
|
Domain Architecture |
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 1 (Potassium channel, inwardly rectifying subfamily J member 1) (ATP-regulated potassium channel ROM-K) (Kir1.1). | |||||
|
IRK1_HUMAN
|
||||||
| θ value | 1.22964e-73 (rank : 23) | NC score | 0.985055 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48048, Q6LD67 | Gene names | KCNJ1, ROMK1 | |||
|
Domain Architecture |
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 1 (Potassium channel, inwardly rectifying subfamily J member 1) (ATP-regulated potassium channel ROM-K) (Kir1.1). | |||||
|
IRK16_HUMAN
|
||||||
| θ value | 3.34864e-71 (rank : 24) | NC score | 0.983480 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NPI9 | Gene names | KCNJ16 | |||
|
Domain Architecture |
|
|||||
| Description | Inward rectifier potassium channel 16 (Potassium channel, inwardly rectifying subfamily J member 16) (Inward rectifier K(+) channel Kir5.1). | |||||
|
IRK16_MOUSE
|
||||||
| θ value | 2.17053e-70 (rank : 25) | NC score | 0.983456 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z307 | Gene names | Kcnj16 | |||
|
Domain Architecture |
|
|||||
| Description | Inward rectifier potassium channel 16 (Potassium channel, inwardly rectifying subfamily J member 16) (Inward rectifier K(+) channel Kir5.1). | |||||
|
IRK10_MOUSE
|
||||||
| θ value | 1.31987e-59 (rank : 26) | NC score | 0.976070 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JM63 | Gene names | Kcnj10 | |||
|
Domain Architecture |
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 10 (Potassium channel, inwardly rectifying subfamily J member 10) (Inward rectifier K(+) channel Kir4.1). | |||||
|
IRK10_HUMAN
|
||||||
| θ value | 2.94036e-59 (rank : 27) | NC score | 0.975848 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78508, Q8N4I7, Q92808 | Gene names | KCNJ10 | |||
|
Domain Architecture |
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 10 (Potassium channel, inwardly rectifying subfamily J member 10) (Inward rectifier K(+) channel Kir1.2) (ATP-dependent inwardly rectifying potassium channel Kir4.1). | |||||
|
IRK15_MOUSE
|
||||||
| θ value | 3.974e-56 (rank : 28) | NC score | 0.975351 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88932, Q9JK34 | Gene names | Kcnj15 | |||
|
Domain Architecture |
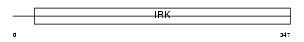
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 15 (Potassium channel, inwardly rectifying subfamily J member 15) (Inward rectifier K(+) channel Kir4.2). | |||||
|
IRK15_HUMAN
|
||||||
| θ value | 5.73848e-55 (rank : 29) | NC score | 0.975116 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99712, O00564, Q96L28, Q99446 | Gene names | KCNJ15, KCNJ14 | |||
|
Domain Architecture |
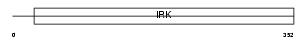
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 15 (Potassium channel, inwardly rectifying subfamily J member 15) (Inward rectifier K(+) channel Kir4.2) (Kir1.3). | |||||
|
IRK13_HUMAN
|
||||||
| θ value | 2.26708e-35 (rank : 30) | NC score | 0.967327 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60928, O76023, Q8N3Y4 | Gene names | KCNJ13 | |||
|
Domain Architecture |
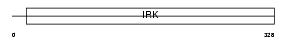
|
|||||
| Description | Inward rectifier potassium channel 13 (Potassium channel, inwardly rectifying subfamily J member 13) (Inward rectifier K(+) channel Kir7.1). | |||||
|
CTGE5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.000526 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
S12A7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.002842 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
PIGC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.011298 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CXR4, Q3U7H3 | Gene names | Pigc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit C (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class C protein) (PIG-C). | |||||
|
POLK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.003977 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUG2, Q7TPY7 | Gene names | Polk, Dinb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase kappa (EC 2.7.7.7) (DINB protein) (DINP). | |||||
|
IRK8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15842, O00657 | Gene names | KCNJ8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 8 (Potassium channel, inwardly rectifying subfamily J member 8) (Inwardly rectifier K(+) channel Kir6.1) (uKATP-1). | |||||
|
IRK8_MOUSE
|
||||||
| NC score | 0.999852 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97794 | Gene names | Kcnj8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 8 (Potassium channel, inwardly rectifying subfamily J member 8) (Inwardly rectifier K(+) channel Kir6.1) (uKATP-1). | |||||
|
IRK11_HUMAN
|
||||||
| NC score | 0.998328 (rank : 3) | θ value | 2.19465e-147 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14654, Q58EX3 | Gene names | KCNJ11 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 11 (Potassium channel, inwardly rectifying subfamily J member 11) (Inward rectifier K(+) channel Kir6.2) (IKATP). | |||||
|
IRK11_MOUSE
|
||||||
| NC score | 0.998286 (rank : 4) | θ value | 4.13891e-146 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61743, Q9QX21 | Gene names | Kcnj11 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 11 (Potassium channel, inwardly rectifying subfamily J member 11) (Inward rectifier K(+) channel Kir6.2). | |||||
|
IRK9_MOUSE
|
||||||
| NC score | 0.990243 (rank : 5) | θ value | 8.47616e-91 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48543, Q9WUE1 | Gene names | Kcnj9, Girk3 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 3 (GIRK3) (Potassium channel, inwardly rectifying subfamily J member 9) (Inwardly rectifier K(+) channel Kir3.3). | |||||
|
IRK9_HUMAN
|
||||||
| NC score | 0.990218 (rank : 6) | θ value | 3.22093e-90 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92806, Q5JW75 | Gene names | KCNJ9, GIRK3 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 3 (GIRK3) (Potassium channel, inwardly rectifying subfamily J member 9) (Inwardly rectifier K(+) channel Kir3.3). | |||||
|
IRK6_HUMAN
|
||||||
| NC score | 0.989366 (rank : 7) | θ value | 5.1304e-96 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48051, Q53WW6 | Gene names | KCNJ6, GIRK2, KATP2, KCNJ7 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 2 (GIRK2) (Potassium channel, inwardly rectifying subfamily J member 6) (Inward rectifier K(+) channel Kir3.2) (KATP-2) (BIR1). | |||||
|
IRK6_MOUSE
|
||||||
| NC score | 0.988865 (rank : 8) | θ value | 6.07444e-89 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48542, O70290, P70216, P70306, P70307, P70308, P70309, P70454, Q9QYH5 | Gene names | Kcnj6, Girk2, Kcnj7, W | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 2 (GIRK2) (Potassium channel, inwardly rectifying subfamily J member 6) (Inward rectifier K(+) channel Kir3.2). | |||||
|
IRK5_MOUSE
|
||||||
| NC score | 0.988777 (rank : 9) | θ value | 1.88829e-90 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48545, P97508 | Gene names | Kcnj5, Girk4 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 4 (GIRK4) (Potassium channel, inwardly rectifying subfamily J member 5) (Inward rectifier K(+) channel Kir3.4) (Heart KATP channel) (KATP-1) (Cardiac inward rectifier) (CIR). | |||||
|
IRK5_HUMAN
|
||||||
| NC score | 0.988765 (rank : 10) | θ value | 2.63489e-92 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48544, Q6DK13, Q6DK14, Q92807 | Gene names | KCNJ5, GIRK4 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 4 (GIRK4) (Potassium channel, inwardly rectifying subfamily J member 5) (Inward rectifier K(+) channel Kir3.4) (Heart KATP channel) (KATP-1) (Cardiac inward rectifier) (CIR). | |||||
|
IRK3_MOUSE
|
||||||
| NC score | 0.988225 (rank : 11) | θ value | 3.22093e-90 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P63250, P35562 | Gene names | Kcnj3, Girk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-activated inward rectifier potassium channel 1 (GIRK1) (Potassium channel, inwardly rectifying subfamily J member 3) (Inward rectifier K(+) channel Kir3.1). | |||||
|
IRK3_HUMAN
|
||||||
| NC score | 0.988195 (rank : 12) | θ value | 4.20666e-90 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48549, Q8TBI0 | Gene names | KCNJ3, GIRK1 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 1 (GIRK1) (Potassium channel, inwardly rectifying subfamily J member 3) (Inward rectifier K(+) channel Kir3.1). | |||||
|
IRK4_HUMAN
|
||||||
| NC score | 0.987986 (rank : 13) | θ value | 9.69798e-87 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48050 | Gene names | KCNJ4 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 4 (Potassium channel, inwardly rectifying subfamily J member 4) (Inward rectifier K(+) channel Kir2.3) (Hippocampal inward rectifier) (HIR) (HRK1) (HIRK2). | |||||
|
IRK4_MOUSE
|
||||||
| NC score | 0.987933 (rank : 14) | θ value | 2.82168e-86 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52189 | Gene names | Kcnj4, Irk3 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 4 (Potassium channel, inwardly rectifying subfamily J member 4) (Inward rectifier K(+) channel Kir2.3) (IRK3). | |||||
|
IRK2_HUMAN
|
||||||
| NC score | 0.986120 (rank : 15) | θ value | 1.00126e-91 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P63252, O15110, P48049 | Gene names | KCNJ2, HIRK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inward rectifier potassium channel 2 (Potassium channel, inwardly rectifying subfamily J member 2) (Inward rectifier K(+) channel Kir2.1) (Cardiac inward rectifier potassium channel) (IRK1). | |||||
|
IRK2_MOUSE
|
||||||
| NC score | 0.985806 (rank : 16) | θ value | 2.91321e-91 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35561 | Gene names | Kcnj2, Irk1 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 2 (Potassium channel, inwardly rectifying subfamily J member 2) (Inward rectifier K(+) channel Kir2.1). | |||||
|
IRK12_HUMAN
|
||||||
| NC score | 0.985703 (rank : 17) | θ value | 1.54831e-84 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14500, O43401, Q8NG63 | Gene names | KCNJ12, IRK2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 12 (Potassium channel, inwardly rectifying subfamily J member 12) (Inward rectifier K(+) channel Kir2.2) (IRK2). | |||||
|
IRK14_HUMAN
|
||||||
| NC score | 0.985697 (rank : 18) | θ value | 2.02684e-76 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UNX9 | Gene names | KCNJ14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-sensitive inward rectifier potassium channel 14 (Potassium channel, inwardly rectifying subfamily J member 14) (Inward rectifier K(+) channel Kir2.4) (IRK4). | |||||
|
IRK1_MOUSE
|
||||||
| NC score | 0.985627 (rank : 19) | θ value | 4.99229e-75 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88335 | Gene names | Kcnj1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 1 (Potassium channel, inwardly rectifying subfamily J member 1) (ATP-regulated potassium channel ROM-K) (Kir1.1). | |||||
|
IRK14_MOUSE
|
||||||
| NC score | 0.985538 (rank : 20) | θ value | 5.8972e-76 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8JZN3, Q8BMK3, Q8BXM0 | Gene names | Kcnj14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-sensitive inward rectifier potassium channel 14 (Potassium channel, inwardly rectifying subfamily J member 14) (Inward rectifier K(+) channel Kir2.4) (IRK4). | |||||
|
IRK12_MOUSE
|
||||||
| NC score | 0.985515 (rank : 21) | θ value | 5.88354e-84 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52187, Q9QYF2 | Gene names | Kcnj12, Irk2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 12 (Potassium channel, inwardly rectifying subfamily J member 12) (Inward rectifier K(+) channel Kir2.2) (IRK2). | |||||
|
IRKI_HUMAN
|
||||||
| NC score | 0.985312 (rank : 22) | θ value | 1.35638e-80 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15756 | Gene names | KCNJN1 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifying K(+) channel negative regulator Kir2.2v. | |||||
|
IRK1_HUMAN
|
||||||
| NC score | 0.985055 (rank : 23) | θ value | 1.22964e-73 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48048, Q6LD67 | Gene names | KCNJ1, ROMK1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 1 (Potassium channel, inwardly rectifying subfamily J member 1) (ATP-regulated potassium channel ROM-K) (Kir1.1). | |||||
|
IRK16_HUMAN
|
||||||
| NC score | 0.983480 (rank : 24) | θ value | 3.34864e-71 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NPI9 | Gene names | KCNJ16 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 16 (Potassium channel, inwardly rectifying subfamily J member 16) (Inward rectifier K(+) channel Kir5.1). | |||||
|
IRK16_MOUSE
|
||||||
| NC score | 0.983456 (rank : 25) | θ value | 2.17053e-70 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z307 | Gene names | Kcnj16 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 16 (Potassium channel, inwardly rectifying subfamily J member 16) (Inward rectifier K(+) channel Kir5.1). | |||||
|
IRK10_MOUSE
|
||||||
| NC score | 0.976070 (rank : 26) | θ value | 1.31987e-59 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JM63 | Gene names | Kcnj10 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 10 (Potassium channel, inwardly rectifying subfamily J member 10) (Inward rectifier K(+) channel Kir4.1). | |||||
|
IRK10_HUMAN
|
||||||
| NC score | 0.975848 (rank : 27) | θ value | 2.94036e-59 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78508, Q8N4I7, Q92808 | Gene names | KCNJ10 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 10 (Potassium channel, inwardly rectifying subfamily J member 10) (Inward rectifier K(+) channel Kir1.2) (ATP-dependent inwardly rectifying potassium channel Kir4.1). | |||||
|
IRK15_MOUSE
|
||||||
| NC score | 0.975351 (rank : 28) | θ value | 3.974e-56 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88932, Q9JK34 | Gene names | Kcnj15 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 15 (Potassium channel, inwardly rectifying subfamily J member 15) (Inward rectifier K(+) channel Kir4.2). | |||||
|
IRK15_HUMAN
|
||||||
| NC score | 0.975116 (rank : 29) | θ value | 5.73848e-55 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99712, O00564, Q96L28, Q99446 | Gene names | KCNJ15, KCNJ14 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 15 (Potassium channel, inwardly rectifying subfamily J member 15) (Inward rectifier K(+) channel Kir4.2) (Kir1.3). | |||||
|
IRK13_HUMAN
|
||||||
| NC score | 0.967327 (rank : 30) | θ value | 2.26708e-35 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60928, O76023, Q8N3Y4 | Gene names | KCNJ13 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 13 (Potassium channel, inwardly rectifying subfamily J member 13) (Inward rectifier K(+) channel Kir7.1). | |||||
|
PIGC_MOUSE
|
||||||
| NC score | 0.011298 (rank : 31) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CXR4, Q3U7H3 | Gene names | Pigc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit C (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class C protein) (PIG-C). | |||||
|
POLK_MOUSE
|
||||||
| NC score | 0.003977 (rank : 32) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUG2, Q7TPY7 | Gene names | Polk, Dinb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase kappa (EC 2.7.7.7) (DINB protein) (DINP). | |||||
|
S12A7_MOUSE
|
||||||
| NC score | 0.002842 (rank : 33) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
CTGE5_HUMAN
|
||||||
| NC score | 0.000526 (rank : 34) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||