Please be patient as the page loads
|
GIMA3_MOUSE
|
||||||
| SwissProt Accessions | Q99MI6, Q4VBX0, Q549W4, Q8CF04 | Gene names | Gimap3, Ian4 | |||
|
Domain Architecture |
|
|||||
| Description | GTPase, IMAP family member 3 (Immunity-associated nucleotide 4 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GIMA3_MOUSE
|
||||||
| θ value | 4.56552e-153 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99MI6, Q4VBX0, Q549W4, Q8CF04 | Gene names | Gimap3, Ian4 | |||
|
Domain Architecture |

|
|||||
| Description | GTPase, IMAP family member 3 (Immunity-associated nucleotide 4 protein). | |||||
|
GIMA5_MOUSE
|
||||||
| θ value | 1.92707e-135 (rank : 2) | NC score | 0.996180 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BWF2, Q501L7, Q549X3 | Gene names | Gimap5, Ian4l1, Imap3 | |||
|
Domain Architecture |

|
|||||
| Description | GTPase, IMAP family member 5 (Immunity-associated nucleotide 4-like 1 protein) (Immunity-associated protein 3). | |||||
|
GIMA5_HUMAN
|
||||||
| θ value | 7.93343e-89 (rank : 3) | NC score | 0.990817 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96F15, Q6IA75, Q96NE4, Q9NUK9 | Gene names | GIMAP5, IAN4L1, IMAP3 | |||
|
Domain Architecture |
|
|||||
| Description | GTPase, IMAP family member 5 (Immunity-associated nucleotide 4-like 1 protein) (Immunity-associated protein 3) (IAN-5). | |||||
|
GIMA1_HUMAN
|
||||||
| θ value | 4.85792e-54 (rank : 4) | NC score | 0.962568 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WWP7, Q8NAZ0 | Gene names | GIMAP1, IMAP1 | |||
|
Domain Architecture |
|
|||||
| Description | GTPase, IMAP family member 1 (Immunity-associated protein 1) (hIMAP1). | |||||
|
GIMA2_HUMAN
|
||||||
| θ value | 1.41344e-53 (rank : 5) | NC score | 0.961485 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UG22, Q96L25 | Gene names | GIMAP2, IMAP2 | |||
|
Domain Architecture |
|
|||||
| Description | GTPase, IMAP family member 2 (Immunity-associated protein 2) (hIMAP2). | |||||
|
GIMA7_HUMAN
|
||||||
| θ value | 5.20228e-48 (rank : 6) | NC score | 0.941022 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHV1 | Gene names | GIMAP7, IAN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase, IMAP family member 7 (Immunity-associated nucleotide 7 protein). | |||||
|
GIMA1_MOUSE
|
||||||
| θ value | 2.58187e-47 (rank : 7) | NC score | 0.956298 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70224 | Gene names | Gimap1, Imap1, Imap38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase, IMAP family member 1 (Immunity-associated protein 1) (Immune- associated protein 38) (IAP38). | |||||
|
GIMA4_HUMAN
|
||||||
| θ value | 1.19932e-44 (rank : 8) | NC score | 0.884453 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NUV9 | Gene names | GIMAP4, IAN1, IMAP4 | |||
|
Domain Architecture |
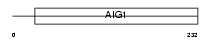
|
|||||
| Description | GTPase, IMAP family member 4 (Immunity-associated protein 4) (Immunity-associated nucleotide 1 protein) (hIAN1). | |||||
|
GIMA4_MOUSE
|
||||||
| θ value | 3.16345e-37 (rank : 9) | NC score | 0.942455 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99JY3 | Gene names | Gimap4, Ian1, Imap4 | |||
|
Domain Architecture |
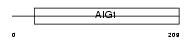
|
|||||
| Description | GTPase, IMAP family member 4 (Immunity-associated protein 4) (Immunity-associated nucleotide 1 protein). | |||||
|
ERAL_HUMAN
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.203874 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75616, O75617, Q8WUY4, Q96LE2, Q96TC0 | Gene names | ERAL1, HERA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (hERA) (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
ERAL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.187884 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CZU4, Q6NV78, Q925U1 | Gene names | Eral1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
SEPT2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.034117 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P42208, Q3U9Y5 | Gene names | Sept2, Nedd-5, Nedd5 | |||
|
Domain Architecture |
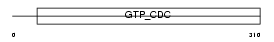
|
|||||
| Description | Septin-2 (Protein NEDD5) (Neural precursor cell expressed developmentally down-regulated protein 5). | |||||
|
SEPT2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.030199 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15019, Q14132, Q8IUK9, Q96CB0 | Gene names | SEPT2, DIFF6, KIAA0158, NEDD5 | |||
|
Domain Architecture |
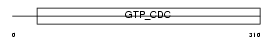
|
|||||
| Description | Septin-2 (Protein NEDD5). | |||||
|
SEPT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.020251 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42209 | Gene names | Sept1, Diff6, Pnutl3 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-1 (Differentiation protein 6) (Diff6 protein). | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.008840 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.011863 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
LOXL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.010436 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58215, Q96RS1 | Gene names | LOXL3, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3). | |||||
|
LOXL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.010464 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z175, Q9JJ39 | Gene names | Loxl3, Lor2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3) (Lysyl oxidase-related protein 2). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.007592 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.006612 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
N4BP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.013743 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C7U1, Q3TDC7, Q7TNR2, Q922W0, Q99J06 | Gene names | N4bp3 | |||
|
Domain Architecture |
|
|||||
| Description | NEDD4-binding protein 3 (N4BP3). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.005864 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RASF8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.006123 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CJ96, Q8CCW4, Q9CU91 | Gene names | Rassf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing protein 8 (Carcinoma-associated protein HOJ-1 homolog). | |||||
|
GIMA3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.56552e-153 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99MI6, Q4VBX0, Q549W4, Q8CF04 | Gene names | Gimap3, Ian4 | |||
|
Domain Architecture |

|
|||||
| Description | GTPase, IMAP family member 3 (Immunity-associated nucleotide 4 protein). | |||||
|
GIMA5_MOUSE
|
||||||
| NC score | 0.996180 (rank : 2) | θ value | 1.92707e-135 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BWF2, Q501L7, Q549X3 | Gene names | Gimap5, Ian4l1, Imap3 | |||
|
Domain Architecture |

|
|||||
| Description | GTPase, IMAP family member 5 (Immunity-associated nucleotide 4-like 1 protein) (Immunity-associated protein 3). | |||||
|
GIMA5_HUMAN
|
||||||
| NC score | 0.990817 (rank : 3) | θ value | 7.93343e-89 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96F15, Q6IA75, Q96NE4, Q9NUK9 | Gene names | GIMAP5, IAN4L1, IMAP3 | |||
|
Domain Architecture |

|
|||||
| Description | GTPase, IMAP family member 5 (Immunity-associated nucleotide 4-like 1 protein) (Immunity-associated protein 3) (IAN-5). | |||||
|
GIMA1_HUMAN
|
||||||
| NC score | 0.962568 (rank : 4) | θ value | 4.85792e-54 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WWP7, Q8NAZ0 | Gene names | GIMAP1, IMAP1 | |||
|
Domain Architecture |

|
|||||
| Description | GTPase, IMAP family member 1 (Immunity-associated protein 1) (hIMAP1). | |||||
|
GIMA2_HUMAN
|
||||||
| NC score | 0.961485 (rank : 5) | θ value | 1.41344e-53 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UG22, Q96L25 | Gene names | GIMAP2, IMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | GTPase, IMAP family member 2 (Immunity-associated protein 2) (hIMAP2). | |||||
|
GIMA1_MOUSE
|
||||||
| NC score | 0.956298 (rank : 6) | θ value | 2.58187e-47 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70224 | Gene names | Gimap1, Imap1, Imap38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase, IMAP family member 1 (Immunity-associated protein 1) (Immune- associated protein 38) (IAP38). | |||||
|
GIMA4_MOUSE
|
||||||
| NC score | 0.942455 (rank : 7) | θ value | 3.16345e-37 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99JY3 | Gene names | Gimap4, Ian1, Imap4 | |||
|
Domain Architecture |

|
|||||
| Description | GTPase, IMAP family member 4 (Immunity-associated protein 4) (Immunity-associated nucleotide 1 protein). | |||||
|
GIMA7_HUMAN
|
||||||
| NC score | 0.941022 (rank : 8) | θ value | 5.20228e-48 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHV1 | Gene names | GIMAP7, IAN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase, IMAP family member 7 (Immunity-associated nucleotide 7 protein). | |||||
|
GIMA4_HUMAN
|
||||||
| NC score | 0.884453 (rank : 9) | θ value | 1.19932e-44 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NUV9 | Gene names | GIMAP4, IAN1, IMAP4 | |||
|
Domain Architecture |

|
|||||
| Description | GTPase, IMAP family member 4 (Immunity-associated protein 4) (Immunity-associated nucleotide 1 protein) (hIAN1). | |||||
|
ERAL_HUMAN
|
||||||
| NC score | 0.203874 (rank : 10) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75616, O75617, Q8WUY4, Q96LE2, Q96TC0 | Gene names | ERAL1, HERA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (hERA) (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
ERAL_MOUSE
|
||||||
| NC score | 0.187884 (rank : 11) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CZU4, Q6NV78, Q925U1 | Gene names | Eral1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
SEPT2_MOUSE
|
||||||
| NC score | 0.034117 (rank : 12) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P42208, Q3U9Y5 | Gene names | Sept2, Nedd-5, Nedd5 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-2 (Protein NEDD5) (Neural precursor cell expressed developmentally down-regulated protein 5). | |||||
|
SEPT2_HUMAN
|
||||||
| NC score | 0.030199 (rank : 13) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15019, Q14132, Q8IUK9, Q96CB0 | Gene names | SEPT2, DIFF6, KIAA0158, NEDD5 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-2 (Protein NEDD5). | |||||
|
SEPT1_MOUSE
|
||||||
| NC score | 0.020251 (rank : 14) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42209 | Gene names | Sept1, Diff6, Pnutl3 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-1 (Differentiation protein 6) (Diff6 protein). | |||||
|
N4BP3_MOUSE
|
||||||
| NC score | 0.013743 (rank : 15) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C7U1, Q3TDC7, Q7TNR2, Q922W0, Q99J06 | Gene names | N4bp3 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 3 (N4BP3). | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.011863 (rank : 16) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
LOXL3_MOUSE
|
||||||
| NC score | 0.010464 (rank : 17) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z175, Q9JJ39 | Gene names | Loxl3, Lor2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3) (Lysyl oxidase-related protein 2). | |||||
|
LOXL3_HUMAN
|
||||||
| NC score | 0.010436 (rank : 18) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58215, Q96RS1 | Gene names | LOXL3, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3). | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.008840 (rank : 19) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.007592 (rank : 20) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.006612 (rank : 21) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
RASF8_MOUSE
|
||||||
| NC score | 0.006123 (rank : 22) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CJ96, Q8CCW4, Q9CU91 | Gene names | Rassf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing protein 8 (Carcinoma-associated protein HOJ-1 homolog). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.005864 (rank : 23) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||