Please be patient as the page loads
|
F262_HUMAN
|
||||||
| SwissProt Accessions | O60825, O60824, Q9H3P1 | Gene names | PFKFB2 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (6PF-2-K/Fru- 2,6-P2ASE heart-type isozyme) (PFK-2/FBPase-2) [Includes: 6- phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
F261_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991632 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16118, Q99951 | Gene names | PFKFB1, F6PK, PFRX | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (6PF-2-K/Fru- 2,6-P2ASE liver isozyme) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
F262_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60825, O60824, Q9H3P1 | Gene names | PFKFB2 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (6PF-2-K/Fru- 2,6-P2ASE heart-type isozyme) (PFK-2/FBPase-2) [Includes: 6- phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
F262_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999054 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70265, Q8VEI9 | Gene names | Pfkfb2 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (6PF-2-K/Fru- 2,6-P2ASE heart-type isozyme) (PFK-2/FBPase-2) [Includes: 6- phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
F263_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.991669 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16875, O43622, O75902 | Gene names | PFKFB3 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (6PF-2-K/Fru- 2,6-P2ASE brain/placenta-type isozyme) (iPFK-2) (NY-REN-56 antigen) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6- bisphosphatase (EC 3.1.3.46)]. | |||||
|
F264_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.990773 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16877 | Gene names | PFKFB4 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (6PF-2-K/Fru- 2,6-P2ASE testis-type isozyme) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
CN37_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 6) | NC score | 0.112620 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P09543 | Gene names | CNP | |||
|
Domain Architecture |
|
|||||
| Description | 2',3'-cyclic-nucleotide 3'-phosphodiesterase (EC 3.1.4.37) (CNP) (CNPase). | |||||
|
PGAM2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 7) | NC score | 0.084622 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15259 | Gene names | PGAM2, PGAMM | |||
|
Domain Architecture |
|
|||||
| Description | Phosphoglycerate mutase 2 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme M) (PGAM-M) (BPG-dependent PGAM 2) (Muscle-specific phosphoglycerate mutase). | |||||
|
CN37_MOUSE
|
||||||
| θ value | 0.279714 (rank : 8) | NC score | 0.095213 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16330, Q61424, Q8C7C9, Q91V42, Q923F3 | Gene names | Cnp, Cnp1 | |||
|
Domain Architecture |
|
|||||
| Description | 2',3'-cyclic-nucleotide 3'-phosphodiesterase (EC 3.1.4.37) (CNP) (CNPase). | |||||
|
PGAM2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.086679 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70250 | Gene names | Pgam2 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphoglycerate mutase 2 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme M) (PGAM-M) (BPG-dependent PGAM 2) (Muscle-specific phosphoglycerate mutase). | |||||
|
ARF6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.022452 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62330, P26438 | Gene names | ARF6 | |||
|
Domain Architecture |
|
|||||
| Description | ADP-ribosylation factor 6. | |||||
|
ARF6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.022452 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62331, P26438 | Gene names | Arf6 | |||
|
Domain Architecture |
|
|||||
| Description | ADP-ribosylation factor 6. | |||||
|
EXOC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.038615 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KP1, Q5JPC8, Q96AN6, Q9NUZ8, Q9UJM7 | Gene names | EXOC2, SEC5, SEC5L1 | |||
|
Domain Architecture |
|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
PGAM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.087690 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18669, Q9BWC0 | Gene names | PGAM1 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1). | |||||
|
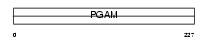
PGAM1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.087670 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBJ1, Q9D6W0, Q9ERT3 | Gene names | Pgam1 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1). | |||||
|
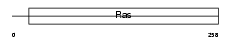
XAB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.032813 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCE2 | Gene names | Xab1, Mbdin | |||
|
Domain Architecture |
|
|||||
| Description | XPA-binding protein 1. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.028562 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.028962 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
SEM6A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.006817 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
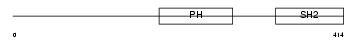
LNK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.008877 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09039 | Gene names | Lnk | |||
|
Domain Architecture |
|
|||||
| Description | Lymphocyte-specific adapter protein Lnk (Signal transduction protein Lnk) (Lymphocyte adapter protein). | |||||
|
ORCT4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.011172 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y267, Q6DJT3 | Gene names | SLC22A14, ORCTL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Organic-cation transporter-like 4 (Solute carrier family 22 member 14). | |||||
|
EPIPL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.006851 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58107, Q76E58 | Gene names | EPPK1, EPIPL | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin (450 kDa epidermal antigen). | |||||
|
SEM6A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.006044 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
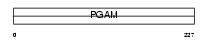
PGAM4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.055276 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N0Y7, Q8NI24, Q8NI25, Q8NI26 | Gene names | PGAM4, PGAM3 | |||
|
Domain Architecture |
|
|||||
| Description | Probable phosphoglycerate mutase 4 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13). | |||||
|
F262_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60825, O60824, Q9H3P1 | Gene names | PFKFB2 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (6PF-2-K/Fru- 2,6-P2ASE heart-type isozyme) (PFK-2/FBPase-2) [Includes: 6- phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
F262_MOUSE
|
||||||
| NC score | 0.999054 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70265, Q8VEI9 | Gene names | Pfkfb2 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (6PF-2-K/Fru- 2,6-P2ASE heart-type isozyme) (PFK-2/FBPase-2) [Includes: 6- phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
F263_HUMAN
|
||||||
| NC score | 0.991669 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16875, O43622, O75902 | Gene names | PFKFB3 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (6PF-2-K/Fru- 2,6-P2ASE brain/placenta-type isozyme) (iPFK-2) (NY-REN-56 antigen) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6- bisphosphatase (EC 3.1.3.46)]. | |||||
|
F261_HUMAN
|
||||||
| NC score | 0.991632 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16118, Q99951 | Gene names | PFKFB1, F6PK, PFRX | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (6PF-2-K/Fru- 2,6-P2ASE liver isozyme) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
F264_HUMAN
|
||||||
| NC score | 0.990773 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16877 | Gene names | PFKFB4 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (6PF-2-K/Fru- 2,6-P2ASE testis-type isozyme) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
CN37_HUMAN
|
||||||
| NC score | 0.112620 (rank : 6) | θ value | 0.0431538 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P09543 | Gene names | CNP | |||
|
Domain Architecture |

|
|||||
| Description | 2',3'-cyclic-nucleotide 3'-phosphodiesterase (EC 3.1.4.37) (CNP) (CNPase). | |||||
|
CN37_MOUSE
|
||||||
| NC score | 0.095213 (rank : 7) | θ value | 0.279714 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16330, Q61424, Q8C7C9, Q91V42, Q923F3 | Gene names | Cnp, Cnp1 | |||
|
Domain Architecture |

|
|||||
| Description | 2',3'-cyclic-nucleotide 3'-phosphodiesterase (EC 3.1.4.37) (CNP) (CNPase). | |||||
|
PGAM1_HUMAN
|
||||||
| NC score | 0.087690 (rank : 8) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18669, Q9BWC0 | Gene names | PGAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1). | |||||
|
PGAM1_MOUSE
|
||||||
| NC score | 0.087670 (rank : 9) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBJ1, Q9D6W0, Q9ERT3 | Gene names | Pgam1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1). | |||||
|
PGAM2_MOUSE
|
||||||
| NC score | 0.086679 (rank : 10) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70250 | Gene names | Pgam2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoglycerate mutase 2 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme M) (PGAM-M) (BPG-dependent PGAM 2) (Muscle-specific phosphoglycerate mutase). | |||||
|
PGAM2_HUMAN
|
||||||
| NC score | 0.084622 (rank : 11) | θ value | 0.21417 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15259 | Gene names | PGAM2, PGAMM | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoglycerate mutase 2 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme M) (PGAM-M) (BPG-dependent PGAM 2) (Muscle-specific phosphoglycerate mutase). | |||||
|
PGAM4_HUMAN
|
||||||
| NC score | 0.055276 (rank : 12) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N0Y7, Q8NI24, Q8NI25, Q8NI26 | Gene names | PGAM4, PGAM3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phosphoglycerate mutase 4 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13). | |||||
|
EXOC2_HUMAN
|
||||||
| NC score | 0.038615 (rank : 13) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KP1, Q5JPC8, Q96AN6, Q9NUZ8, Q9UJM7 | Gene names | EXOC2, SEC5, SEC5L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
XAB1_MOUSE
|
||||||
| NC score | 0.032813 (rank : 14) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCE2 | Gene names | Xab1, Mbdin | |||
|
Domain Architecture |

|
|||||
| Description | XPA-binding protein 1. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.028962 (rank : 15) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.028562 (rank : 16) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ARF6_HUMAN
|
||||||
| NC score | 0.022452 (rank : 17) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62330, P26438 | Gene names | ARF6 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor 6. | |||||
|
ARF6_MOUSE
|
||||||
| NC score | 0.022452 (rank : 18) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62331, P26438 | Gene names | Arf6 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor 6. | |||||
|
ORCT4_HUMAN
|
||||||
| NC score | 0.011172 (rank : 19) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y267, Q6DJT3 | Gene names | SLC22A14, ORCTL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Organic-cation transporter-like 4 (Solute carrier family 22 member 14). | |||||
|
LNK_MOUSE
|
||||||
| NC score | 0.008877 (rank : 20) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09039 | Gene names | Lnk | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific adapter protein Lnk (Signal transduction protein Lnk) (Lymphocyte adapter protein). | |||||
|
EPIPL_HUMAN
|
||||||
| NC score | 0.006851 (rank : 21) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58107, Q76E58 | Gene names | EPPK1, EPIPL | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin (450 kDa epidermal antigen). | |||||
|
SEM6A_HUMAN
|
||||||
| NC score | 0.006817 (rank : 22) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM6A_MOUSE
|
||||||
| NC score | 0.006044 (rank : 23) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||