Please be patient as the page loads
|
DPOE3_MOUSE
|
||||||
| SwissProt Accessions | Q9JKP7, Q8R198 | Gene names | Pole3 | |||
|
Domain Architecture |
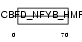
|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (YB-like protein 1) (YBL1) (NF-YB-like protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DPOE3_MOUSE
|
||||||
| θ value | 2.18568e-46 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JKP7, Q8R198 | Gene names | Pole3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (YB-like protein 1) (YBL1) (NF-YB-like protein). | |||||
|
DPOE3_HUMAN
|
||||||
| θ value | 8.30557e-46 (rank : 2) | NC score | 0.977565 (rank : 2) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NRF9, Q5W0U1, Q8N758, Q8NCE5, Q9NR32 | Gene names | POLE3, CHRAC17 | |||
|
Domain Architecture |
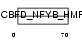
|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (Chromatin accessibility complex 17) (HuCHRAC17) (CHRAC-17) (Arsenic- transactivated protein) (AsTP). | |||||
|
NFYB_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 3) | NC score | 0.719323 (rank : 3) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P25208, Q96IY8 | Gene names | NFYB, HAP3 | |||
|
Domain Architecture |
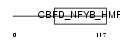
|
|||||
| Description | Nuclear transcription factor Y subunit beta (Nuclear transcription factor Y subunit B) (NF-YB) (CAAT-box DNA-binding protein subunit B). | |||||
|
NFYB_MOUSE
|
||||||
| θ value | 9.26847e-13 (rank : 4) | NC score | 0.719279 (rank : 4) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P63139, P22569, Q3UK54 | Gene names | Nfyb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear transcription factor Y subunit beta (Nuclear transcription factor Y subunit B) (NF-YB) (CAAT-box DNA-binding protein subunit B). | |||||
|
TBAP_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 5) | NC score | 0.676723 (rank : 5) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01658 | Gene names | DR1 | |||
|
Domain Architecture |
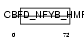
|
|||||
| Description | TATA-binding protein-associated phosphoprotein (Down-regulator of transcription 1) (Dr1 protein) (Negative cofactor 2 beta) (NC2 beta). | |||||
|
TBAP_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 6) | NC score | 0.674547 (rank : 6) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91WV0, Q3UT14 | Gene names | Dr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA-binding protein-associated phosphoprotein (Down-regulator of transcription 1) (Negative cofactor 2 beta) (NC2 beta). | |||||
|
CCD16_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 7) | NC score | 0.215492 (rank : 10) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
CCD16_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 8) | NC score | 0.213792 (rank : 11) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
CHRC1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 9) | NC score | 0.227212 (rank : 9) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JKP8, Q91VG2 | Gene names | Chrac1 | |||
|
Domain Architecture |
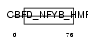
|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (DNA polymerase epsilon subunit p15) (YC-like protein 1) (YCL1) (NF-YC-like protein). | |||||
|
DPOE4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.232923 (rank : 8) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NR33, Q53TR2 | Gene names | POLE4 | |||
|
Domain Architecture |
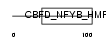
|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
DPOE4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.233692 (rank : 7) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CQ36, Q3TJ13 | Gene names | Pole4 | |||
|
Domain Architecture |
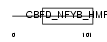
|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.052098 (rank : 31) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.062859 (rank : 23) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.084115 (rank : 17) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.074586 (rank : 21) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
PSMD2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.081275 (rank : 18) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDM4 | Gene names | Psmd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.040217 (rank : 39) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.049322 (rank : 35) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.088768 (rank : 15) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.055617 (rank : 26) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
NFYC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.145381 (rank : 13) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13952, Q5T6K8, Q5T6L1, Q5TZR6, Q92869, Q9HBX1, Q9NXB5, Q9UM67, Q9UML0, Q9UMT7 | Gene names | NFYC | |||
|
Domain Architecture |
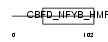
|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C) (Transactivator HSM-1/2). | |||||
|
NFYC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.158676 (rank : 12) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70353, O35088, Q6GU21 | Gene names | Nfyc | |||
|
Domain Architecture |
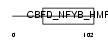
|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.087974 (rank : 16) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.060951 (rank : 24) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
PSMD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.076572 (rank : 20) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13200, Q12932, Q15321, Q96I12 | Gene names | PSMD2, TRAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97) (Tumor necrosis factor type 1 receptor- associated protein 2) (55.11 protein). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.080203 (rank : 19) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.052633 (rank : 30) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.051686 (rank : 33) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.045057 (rank : 37) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.052775 (rank : 29) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.036366 (rank : 43) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.035061 (rank : 47) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
MOR2A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.022118 (rank : 53) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69ZX6, Q5QNQ7, Q6P547 | Gene names | Morc2a, Kiaa0852, Zcwcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2A (Zinc finger CW-type coiled-coil domain protein 1). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.037012 (rank : 42) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.035866 (rank : 45) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
IDLC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.052927 (rank : 28) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14645, Q5TGH0, Q7L0I5 | Gene names | DNALI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axonemal dynein light intermediate polypeptide 1 (Inner dynein arm light chain, axonemal) (hp28). | |||||
|
IDLC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.055859 (rank : 25) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BVN8 | Gene names | Dnali1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axonemal dynein light intermediate polypeptide 1 (Inner dynein arm light chain, axonemal). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.037189 (rank : 41) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.035871 (rank : 44) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
P52K_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.035622 (rank : 46) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.050528 (rank : 34) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
DOCK9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.013248 (rank : 55) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZ29, Q5JUD4, Q5JUD6, Q5T2Q1, Q5TAN8, Q9BZ25, Q9BZ26, Q9BZ27, Q9BZ28, Q9UPU4 | Gene names | DOCK9, KIAA1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
F10C1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.026521 (rank : 51) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BP78 | Gene names | Fra10ac1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FRA10AC1 homolog. | |||||
|
KIF3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.026311 (rank : 52) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
KIF3A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.027016 (rank : 50) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
NAF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.039336 (rank : 40) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.020437 (rank : 54) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.040955 (rank : 38) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.028615 (rank : 49) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.048143 (rank : 36) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.034837 (rank : 48) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
CHRC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.112083 (rank : 14) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRG0 | Gene names | CHRAC1, CHRAC15 | |||
|
Domain Architecture |
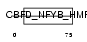
|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (CHRAC-15) (HuCHRAC15) (DNA polymerase epsilon subunit p15). | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.070799 (rank : 22) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.054500 (rank : 27) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.051934 (rank : 32) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
DPOE3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.18568e-46 (rank : 1) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JKP7, Q8R198 | Gene names | Pole3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (YB-like protein 1) (YBL1) (NF-YB-like protein). | |||||
|
DPOE3_HUMAN
|
||||||
| NC score | 0.977565 (rank : 2) | θ value | 8.30557e-46 (rank : 2) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NRF9, Q5W0U1, Q8N758, Q8NCE5, Q9NR32 | Gene names | POLE3, CHRAC17 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (Chromatin accessibility complex 17) (HuCHRAC17) (CHRAC-17) (Arsenic- transactivated protein) (AsTP). | |||||
|
NFYB_HUMAN
|
||||||
| NC score | 0.719323 (rank : 3) | θ value | 9.26847e-13 (rank : 3) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P25208, Q96IY8 | Gene names | NFYB, HAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit beta (Nuclear transcription factor Y subunit B) (NF-YB) (CAAT-box DNA-binding protein subunit B). | |||||
|
NFYB_MOUSE
|
||||||
| NC score | 0.719279 (rank : 4) | θ value | 9.26847e-13 (rank : 4) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P63139, P22569, Q3UK54 | Gene names | Nfyb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear transcription factor Y subunit beta (Nuclear transcription factor Y subunit B) (NF-YB) (CAAT-box DNA-binding protein subunit B). | |||||
|
TBAP_HUMAN
|
||||||
| NC score | 0.676723 (rank : 5) | θ value | 6.21693e-09 (rank : 5) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01658 | Gene names | DR1 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated phosphoprotein (Down-regulator of transcription 1) (Dr1 protein) (Negative cofactor 2 beta) (NC2 beta). | |||||
|
TBAP_MOUSE
|
||||||
| NC score | 0.674547 (rank : 6) | θ value | 6.21693e-09 (rank : 6) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91WV0, Q3UT14 | Gene names | Dr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA-binding protein-associated phosphoprotein (Down-regulator of transcription 1) (Negative cofactor 2 beta) (NC2 beta). | |||||
|
DPOE4_MOUSE
|
||||||
| NC score | 0.233692 (rank : 7) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CQ36, Q3TJ13 | Gene names | Pole4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
DPOE4_HUMAN
|
||||||
| NC score | 0.232923 (rank : 8) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NR33, Q53TR2 | Gene names | POLE4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
CHRC1_MOUSE
|
||||||
| NC score | 0.227212 (rank : 9) | θ value | 0.0193708 (rank : 9) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JKP8, Q91VG2 | Gene names | Chrac1 | |||
|
Domain Architecture |
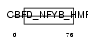
|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (DNA polymerase epsilon subunit p15) (YC-like protein 1) (YCL1) (NF-YC-like protein). | |||||
|
CCD16_MOUSE
|
||||||
| NC score | 0.215492 (rank : 10) | θ value | 0.00665767 (rank : 7) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
CCD16_HUMAN
|
||||||
| NC score | 0.213792 (rank : 11) | θ value | 0.0148317 (rank : 8) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
NFYC_MOUSE
|
||||||
| NC score | 0.158676 (rank : 12) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70353, O35088, Q6GU21 | Gene names | Nfyc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C). | |||||
|
NFYC_HUMAN
|
||||||
| NC score | 0.145381 (rank : 13) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13952, Q5T6K8, Q5T6L1, Q5TZR6, Q92869, Q9HBX1, Q9NXB5, Q9UM67, Q9UML0, Q9UMT7 | Gene names | NFYC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C) (Transactivator HSM-1/2). | |||||
|
CHRC1_HUMAN
|
||||||
| NC score | 0.112083 (rank : 14) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRG0 | Gene names | CHRAC1, CHRAC15 | |||
|
Domain Architecture |

|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (CHRAC-15) (HuCHRAC15) (DNA polymerase epsilon subunit p15). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.088768 (rank : 15) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.087974 (rank : 16) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.084115 (rank : 17) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
PSMD2_MOUSE
|
||||||
| NC score | 0.081275 (rank : 18) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDM4 | Gene names | Psmd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.080203 (rank : 19) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
PSMD2_HUMAN
|
||||||
| NC score | 0.076572 (rank : 20) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13200, Q12932, Q15321, Q96I12 | Gene names | PSMD2, TRAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97) (Tumor necrosis factor type 1 receptor- associated protein 2) (55.11 protein). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.074586 (rank : 21) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.070799 (rank : 22) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.062859 (rank : 23) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.060951 (rank : 24) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
IDLC_MOUSE
|
||||||
| NC score | 0.055859 (rank : 25) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BVN8 | Gene names | Dnali1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axonemal dynein light intermediate polypeptide 1 (Inner dynein arm light chain, axonemal). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.055617 (rank : 26) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.054500 (rank : 27) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
IDLC_HUMAN
|
||||||
| NC score | 0.052927 (rank : 28) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14645, Q5TGH0, Q7L0I5 | Gene names | DNALI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axonemal dynein light intermediate polypeptide 1 (Inner dynein arm light chain, axonemal) (hp28). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.052775 (rank : 29) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.052633 (rank : 30) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.052098 (rank : 31) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.051934 (rank : 32) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.051686 (rank : 33) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.050528 (rank : 34) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.049322 (rank : 35) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.048143 (rank : 36) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.045057 (rank : 37) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.040955 (rank : 38) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.040217 (rank : 39) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.039336 (rank : 40) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.037189 (rank : 41) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.037012 (rank : 42) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.036366 (rank : 43) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.035871 (rank : 44) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.035866 (rank : 45) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.035622 (rank : 46) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.035061 (rank : 47) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.034837 (rank : 48) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.028615 (rank : 49) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
KIF3A_MOUSE
|
||||||
| NC score | 0.027016 (rank : 50) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
F10C1_MOUSE
|
||||||
| NC score | 0.026521 (rank : 51) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BP78 | Gene names | Fra10ac1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FRA10AC1 homolog. | |||||
|
KIF3A_HUMAN
|
||||||
| NC score | 0.026311 (rank : 52) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
MOR2A_MOUSE
|
||||||
| NC score | 0.022118 (rank : 53) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69ZX6, Q5QNQ7, Q6P547 | Gene names | Morc2a, Kiaa0852, Zcwcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2A (Zinc finger CW-type coiled-coil domain protein 1). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.020437 (rank : 54) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
DOCK9_HUMAN
|
||||||
| NC score | 0.013248 (rank : 55) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZ29, Q5JUD4, Q5JUD6, Q5T2Q1, Q5TAN8, Q9BZ25, Q9BZ26, Q9BZ27, Q9BZ28, Q9UPU4 | Gene names | DOCK9, KIAA1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||