Please be patient as the page loads
|
CSP9_MOUSE
|
||||||
| SwissProt Accessions | Q9CZB6, Q3UGV1, Q91VN3, Q9CWV8, Q9D0V9 | Gene names | Crsp9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CSP9_MOUSE
|
||||||
| θ value | 1.06234e-117 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CZB6, Q3UGV1, Q91VN3, Q9CWV8, Q9D0V9 | Gene names | Crsp9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9. | |||||
|
CSP9_HUMAN
|
||||||
| θ value | 7.12574e-114 (rank : 2) | NC score | 0.998778 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43513 | Gene names | CRSP9, ARC34, MED7 | |||
|
Domain Architecture |
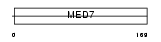
|
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9 (Transcriptional coactivator CRSP33) (RNA polymerase transcriptional regulation mediator subunit 7 homolog) (hMED7) (Activator-recruited cofactor 34 kDa component) (ARC34). | |||||
|
ZYX_MOUSE
|
||||||
| θ value | 1.38821 (rank : 3) | NC score | 0.035145 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62523, P70461 | Gene names | Zyx | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin. | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 4) | NC score | 0.025233 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
SATB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 5) | NC score | 0.049359 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPW6 | Gene names | SATB2, KIAA1034 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB2 (Special AT-rich sequence-binding protein 2). | |||||
|
SATB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 6) | NC score | 0.049355 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VI24 | Gene names | Satb2, Kiaa1034 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB2 (Special AT-rich sequence-binding protein 2). | |||||
|
HDAC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 7) | NC score | 0.014481 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
ZYX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 8) | NC score | 0.024100 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15942 | Gene names | ZYX | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin (Zyxin-2). | |||||
|
EGR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 9) | NC score | 0.004804 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q06889 | Gene names | EGR3, PILOT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 3 (EGR-3) (Zinc finger protein pilot). | |||||
|
EGR3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 10) | NC score | 0.004824 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43300, Q9R276 | Gene names | Egr3, Egr-3 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 3 (EGR-3). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 11) | NC score | 0.019882 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
LRC17_MOUSE
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.009567 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CXD9 | Gene names | Lrrc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 17 precursor. | |||||
|
CSP9_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.06234e-117 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CZB6, Q3UGV1, Q91VN3, Q9CWV8, Q9D0V9 | Gene names | Crsp9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9. | |||||
|
CSP9_HUMAN
|
||||||
| NC score | 0.998778 (rank : 2) | θ value | 7.12574e-114 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43513 | Gene names | CRSP9, ARC34, MED7 | |||
|
Domain Architecture |

|
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9 (Transcriptional coactivator CRSP33) (RNA polymerase transcriptional regulation mediator subunit 7 homolog) (hMED7) (Activator-recruited cofactor 34 kDa component) (ARC34). | |||||
|
SATB2_HUMAN
|
||||||
| NC score | 0.049359 (rank : 3) | θ value | 3.0926 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPW6 | Gene names | SATB2, KIAA1034 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB2 (Special AT-rich sequence-binding protein 2). | |||||
|
SATB2_MOUSE
|
||||||
| NC score | 0.049355 (rank : 4) | θ value | 3.0926 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VI24 | Gene names | Satb2, Kiaa1034 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB2 (Special AT-rich sequence-binding protein 2). | |||||
|
ZYX_MOUSE
|
||||||
| NC score | 0.035145 (rank : 5) | θ value | 1.38821 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62523, P70461 | Gene names | Zyx | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin. | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.025233 (rank : 6) | θ value | 2.36792 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ZYX_HUMAN
|
||||||
| NC score | 0.024100 (rank : 7) | θ value | 6.88961 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15942 | Gene names | ZYX | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin (Zyxin-2). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.019882 (rank : 8) | θ value | 8.99809 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
HDAC4_HUMAN
|
||||||
| NC score | 0.014481 (rank : 9) | θ value | 4.03905 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
LRC17_MOUSE
|
||||||
| NC score | 0.009567 (rank : 10) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CXD9 | Gene names | Lrrc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 17 precursor. | |||||
|
EGR3_MOUSE
|
||||||
| NC score | 0.004824 (rank : 11) | θ value | 8.99809 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43300, Q9R276 | Gene names | Egr3, Egr-3 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 3 (EGR-3). | |||||
|
EGR3_HUMAN
|
||||||
| NC score | 0.004804 (rank : 12) | θ value | 8.99809 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q06889 | Gene names | EGR3, PILOT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 3 (EGR-3) (Zinc finger protein pilot). | |||||