Please be patient as the page loads
|
CP4Z1_HUMAN
|
||||||
| SwissProt Accessions | Q86W10 | Gene names | CYP4Z1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4Z1 (EC 1.14.14.1) (CYPIVZ1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CP4Z1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q86W10 | Gene names | CYP4Z1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4Z1 (EC 1.14.14.1) (CYPIVZ1). | |||||
|
CP4X1_HUMAN
|
||||||
| θ value | 1.91371e-159 (rank : 2) | NC score | 0.995597 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q8N118 | Gene names | CYP4X1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4X1 (EC 1.14.14.1) (CYPIVX1). | |||||
|
CP4AB_HUMAN
|
||||||
| θ value | 8.33966e-147 (rank : 3) | NC score | 0.993024 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q02928, Q06766, Q16865, Q16866, Q8IWY5 | Gene names | CYP4A11 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4A11 precursor (EC 1.14.15.3) (CYPIVA11) (Fatty acid omega-hydroxylase) (P-450 HK omega) (Lauric acid omega-hydroxylase) (CYP4AII) (P450-HL-omega). | |||||
|
CP4AA_MOUSE
|
||||||
| θ value | 7.05993e-146 (rank : 4) | NC score | 0.993311 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O88833, Q9DCS5 | Gene names | Cyp4a10, Cyp4a-10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4A10 (EC 1.14.15.3) (CYPIVA10) (Lauric acid omega- hydroxylase) (P450-LA-omega 1) (P452). | |||||
|
CP4AC_MOUSE
|
||||||
| θ value | 5.05951e-144 (rank : 5) | NC score | 0.992635 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q91WL5, Q3UNE4, Q6P931, Q8N7N3 | Gene names | Cyp4a12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4A12 (EC 1.14.14.1) (CYPIVA12). | |||||
|
CP4AE_MOUSE
|
||||||
| θ value | 1.98959e-140 (rank : 6) | NC score | 0.993840 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O35728, Q3UER2, Q5EBH0 | Gene names | Cyp4a14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4A14 precursor (EC 1.14.15.3) (CYPIVA14) (Lauric acid omega-hydroxylase 3) (P450-LA-omega 3). | |||||
|
CP4B1_MOUSE
|
||||||
| θ value | 4.74654e-134 (rank : 7) | NC score | 0.990630 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q64462 | Gene names | Cyp4b1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4B1 (EC 1.14.14.1) (CYPIVB1). | |||||
|
CP4B1_HUMAN
|
||||||
| θ value | 1.47893e-127 (rank : 8) | NC score | 0.990581 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P13584, Q8TD85, Q8WWU9, Q8WWV0 | Gene names | CYP4B1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4B1 (EC 1.14.14.1) (CYPIVB1) (P450-HP). | |||||
|
CP4F2_HUMAN
|
||||||
| θ value | 7.89671e-105 (rank : 9) | NC score | 0.984303 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P78329, Q16677, Q6NWT4, Q6NWT6, Q9NNZ0, Q9UIU8 | Gene names | CYP4F2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4F2 (EC 1.14.13.30) (CYPIVF2) (Leukotriene-B(4) omega- hydroxylase) (Leukotriene-B(4) 20-monooxygenase) (Cytochrome P450-LTB- omega). | |||||
|
CP4F8_HUMAN
|
||||||
| θ value | 6.68496e-104 (rank : 10) | NC score | 0.985073 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P98187 | Gene names | CYP4F8 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4F8 (EC 1.14.14.1) (CYPIVF8). | |||||
|
CP4FB_HUMAN
|
||||||
| θ value | 1.48926e-103 (rank : 11) | NC score | 0.984599 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9HBI6, O75254, Q96AQ5 | Gene names | CYP4F11 | |||
|
Domain Architecture |
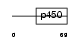
|
|||||
| Description | Cytochrome P450 4F11 (EC 1.14.14.1) (CYPIVF11). | |||||
|
CP4FC_HUMAN
|
||||||
| θ value | 4.33309e-103 (rank : 12) | NC score | 0.985092 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9HCS2, Q9HCS1 | Gene names | CYP4F12 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4F12 (EC 1.14.14.1) (CYPIVF12). | |||||
|
CP4FE_MOUSE
|
||||||
| θ value | 4.33309e-103 (rank : 13) | NC score | 0.982860 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9EP75, Q52L93 | Gene names | Cyp4f14 | |||
|
Domain Architecture |
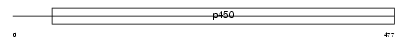
|
|||||
| Description | Cytochrome P450 4F14 (EC 1.14.13.30) (Leukotriene-B4 omega- hydroxylase) (Leukotriene-B4 20-monooxygenase) (Cytochrome P450-LTB- omega) (Cyp4f-14). | |||||
|
CP4F3_HUMAN
|
||||||
| θ value | 3.4333e-100 (rank : 14) | NC score | 0.983776 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q08477, O60634, Q5U740 | Gene names | CYP4F3, LTB4H | |||
|
Domain Architecture |
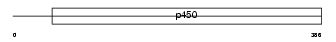
|
|||||
| Description | Cytochrome P450 4F3 (EC 1.14.13.30) (CYPIVF3) (Leukotriene-B(4) omega- hydroxylase) (Leukotriene-B(4) 20-monooxygenase) (Cytochrome P450-LTB- omega). | |||||
|
CP4F3_MOUSE
|
||||||
| θ value | 6.47494e-99 (rank : 15) | NC score | 0.981424 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q99N16, Q9D8N4 | Gene names | Cyp4f3, Cyp4f18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4F3 (EC 1.14.13.30) (CYPIVF3) (Leukotriene-B(4) omega- hydroxylase) (Leukotriene-B(4) 20-monooxygenase) (Cytochrome P450-LTB- omega). | |||||
|
CP4V3_MOUSE
|
||||||
| θ value | 1.84173e-61 (rank : 16) | NC score | 0.968322 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9DBW0 | Gene names | Cyp4v3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4V3 (EC 1.14.-.-). | |||||
|
CP4V2_HUMAN
|
||||||
| θ value | 1.31987e-59 (rank : 17) | NC score | 0.968523 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q6ZWL3, Q6ZTM4 | Gene names | CYP4V2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4V2 (EC 1.14.-.-). | |||||
|
CP343_HUMAN
|
||||||
| θ value | 1.01529e-43 (rank : 18) | NC score | 0.915394 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9HB55, Q9HB52, Q9HB53, Q9HB54, Q9HB57 | Gene names | CYP3A43 | |||
|
Domain Architecture |
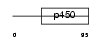
|
|||||
| Description | Cytochrome P450 3A43 (EC 1.14.14.1). | |||||
|
CP3A7_HUMAN
|
||||||
| θ value | 1.91475e-42 (rank : 19) | NC score | 0.912831 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P24462 | Gene names | CYP3A7 | |||
|
Domain Architecture |
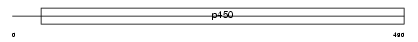
|
|||||
| Description | Cytochrome P450 3A7 (EC 1.14.14.1) (CYPIIIA7) (P450-HFLA). | |||||
|
CP3AP_MOUSE
|
||||||
| θ value | 3.26607e-42 (rank : 20) | NC score | 0.916368 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O09158 | Gene names | Cyp3a25 | |||
|
Domain Architecture |
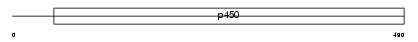
|
|||||
| Description | Cytochrome P450 3A25 (EC 1.14.14.1) (CYPIIIA25). | |||||
|
CP3AD_MOUSE
|
||||||
| θ value | 9.50281e-42 (rank : 21) | NC score | 0.916699 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q64464 | Gene names | Cyp3a13, Cyp3a-13 | |||
|
Domain Architecture |
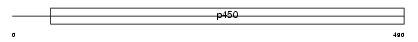
|
|||||
| Description | Cytochrome P450 3A13 (EC 1.14.14.1) (CYPIIIA13). | |||||
|
CP3A4_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 22) | NC score | 0.911627 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P08684, Q16757, Q9UK50 | Gene names | CYP3A4 | |||
|
Domain Architecture |
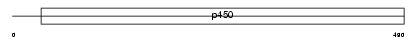
|
|||||
| Description | Cytochrome P450 3A4 (EC 1.14.13.67) (Quinine 3-monooxygenase) (CYPIIIA4) (Nifedipine oxidase) (Taurochenodeoxycholate 6-alpha- hydroxylase) (EC 1.14.13.97) (NF-25) (P450-PCN1). | |||||
|
CP3A3_HUMAN
|
||||||
| θ value | 8.04462e-41 (rank : 23) | NC score | 0.909872 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P05184 | Gene names | CYP3A3 | |||
|
Domain Architecture |
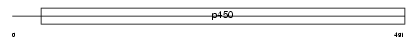
|
|||||
| Description | Cytochrome P450 3A3 (EC 1.14.14.1) (CYPIIIA3) (HLp). | |||||
|
CP341_MOUSE
|
||||||
| θ value | 8.89434e-40 (rank : 24) | NC score | 0.910738 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9JMA7 | Gene names | Cyp3a41 | |||
|
Domain Architecture |
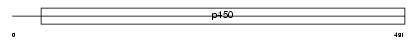
|
|||||
| Description | Cytochrome P450 3A41 (EC 1.14.14.1). | |||||
|
CP3AB_MOUSE
|
||||||
| θ value | 3.37985e-39 (rank : 25) | NC score | 0.909548 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q64459 | Gene names | Cyp3a11, Cyp3a-11 | |||
|
Domain Architecture |
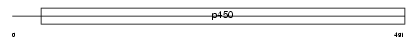
|
|||||
| Description | Cytochrome P450 3A11 (EC 1.14.14.1) (CYPIIIA11) (P-450IIIAM1) (P- 450UT). | |||||
|
CP3AG_MOUSE
|
||||||
| θ value | 1.6774e-38 (rank : 26) | NC score | 0.911589 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q64481 | Gene names | Cyp3a16, Cyp3a-16 | |||
|
Domain Architecture |
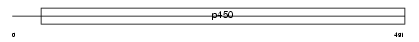
|
|||||
| Description | Cytochrome P450 3A16 (EC 1.14.14.1) (CYPIIIA16). | |||||
|
CP3A5_HUMAN
|
||||||
| θ value | 3.73686e-38 (rank : 27) | NC score | 0.909956 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P20815, Q53WY8, Q75MV0, Q9HB56 | Gene names | CYP3A5 | |||
|
Domain Architecture |
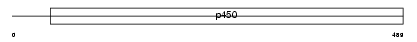
|
|||||
| Description | Cytochrome P450 3A5 (EC 1.14.14.1) (CYPIIIA5) (P450-PCN3) (HLp2). | |||||
|
THAS_MOUSE
|
||||||
| θ value | 4.89182e-30 (rank : 28) | NC score | 0.915290 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P36423, P97505 | Gene names | Tbxas1, Cyp5, Cyp5a1 | |||
|
Domain Architecture |

|
|||||
| Description | Thromboxane-A synthase (EC 5.3.99.5) (TXA synthase) (TXS) (TS) (Cytochrome P450 5A1). | |||||
|
CP46A_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 29) | NC score | 0.924896 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9WVK8 | Gene names | Cyp46a1, Cyp46 | |||
|
Domain Architecture |
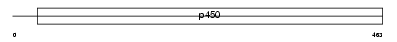
|
|||||
| Description | Cytochrome p450 46A1 (EC 1.14.13.98) (Cholesterol 24-hydroxylase) (CH24H). | |||||
|
CP46A_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 30) | NC score | 0.923007 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9Y6A2 | Gene names | CYP46A1, CYP46 | |||
|
Domain Architecture |
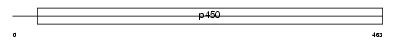
|
|||||
| Description | Cytochrome p450 46A1 (EC 1.14.13.98) (Cholesterol 24-hydroxylase) (CH24H). | |||||
|
CP2J2_HUMAN
|
||||||
| θ value | 8.08199e-25 (rank : 31) | NC score | 0.799806 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P51589 | Gene names | CYP2J2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2J2 (EC 1.14.14.1) (CYPIIJ2) (Arachidonic acid epoxygenase). | |||||
|
CP2R1_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 32) | NC score | 0.815875 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q6VVX0, Q5RT65 | Gene names | CYP2R1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2R1 (EC 1.14.14.-) (Vitamin D 25-hydroxylase). | |||||
|
THAS_HUMAN
|
||||||
| θ value | 6.84181e-24 (rank : 33) | NC score | 0.903184 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P24557, Q16844, Q8IUN1, Q9GZW4, Q9HD77, Q9HD78, Q9HD79, Q9HD80, Q9HD81, Q9HD82, Q9HD83, Q9HD84 | Gene names | TBXAS1, CYP5, CYP5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Thromboxane-A synthase (EC 5.3.99.5) (TXA synthase) (TXS) (Cytochrome P450 5A1). | |||||
|
CP24A_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 34) | NC score | 0.871528 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q64441 | Gene names | Cyp24a1, Cyp-24, Cyp24 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 24A1, mitochondrial precursor (EC 1.14.-.-) (P450- CC24) (Vitamin D(3) 24-hydroxylase) (1,25-dihydroxyvitamin D(3) 24- hydroxylase) (24-OHase). | |||||
|
CP2J6_MOUSE
|
||||||
| θ value | 9.87957e-23 (rank : 35) | NC score | 0.800522 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O54750 | Gene names | Cyp2j6 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2J6 (EC 1.14.14.1) (CYPIIJ6) (Arachidonic acid epoxygenase). | |||||
|
CP24A_HUMAN
|
||||||
| θ value | 1.29031e-22 (rank : 36) | NC score | 0.873365 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q07973, Q15807 | Gene names | CYP24A1, CYP24 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 24A1, mitochondrial precursor (EC 1.14.-.-) (P450- CC24) (Vitamin D(3) 24-hydroxylase) (1,25-dihydroxyvitamin D(3) 24- hydroxylase) (24-OHase). | |||||
|
CP2J5_MOUSE
|
||||||
| θ value | 4.9032e-22 (rank : 37) | NC score | 0.800081 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O54749 | Gene names | Cyp2j5 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2J5 (EC 1.14.14.1) (CYPIIJ5) (Arachidonic acid epoxygenase). | |||||
|
CP17A_MOUSE
|
||||||
| θ value | 6.40375e-22 (rank : 38) | NC score | 0.832550 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P27786 | Gene names | Cyp17a1, Cyp17 | |||
|
Domain Architecture |
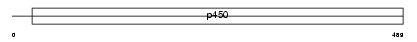
|
|||||
| Description | Cytochrome P450 17A1 (EC 1.14.99.9) (CYPXVII) (P450-C17) (P450c17) (Steroid 17-alpha-hydroxylase/17,20 lyase). | |||||
|
CP27B_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 39) | NC score | 0.872750 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O35084 | Gene names | Cyp27b1, Cyp27b, Cyp40 | |||
|
Domain Architecture |

|
|||||
| Description | 25-hydroxyvitamin D-1 alpha hydroxylase, mitochondrial precursor (EC 1.14.13.13) (Cytochrome P450 subfamily XXVIIB polypeptide 1) (Cytochrome p450 27B1) (Calcidiol 1-monooxygenase) (25-OHD-1 alpha- hydroxylase) (25-hydroxyvitamin D(3) 1-alpha-hydroxylase) (VD3 1A hydroxylase) (P450C1 alpha) (P450VD1-alpha). | |||||
|
CP2R1_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 40) | NC score | 0.811667 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q6VVW9 | Gene names | Cyp2r1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2R1 (EC 1.14.14.-) (Vitamin D 25-hydroxylase). | |||||
|
CP27B_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 41) | NC score | 0.872820 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O15528 | Gene names | CYP27B1, CYP1ALPHA, CYP27B | |||
|
Domain Architecture |

|
|||||
| Description | 25-hydroxyvitamin D-1 alpha hydroxylase, mitochondrial precursor (EC 1.14.13.13) (Cytochrome P450 subfamily XXVIIB polypeptide 1) (Cytochrome p450 27B1) (Calcidiol 1-monooxygenase) (25-OHD-1 alpha- hydroxylase) (25-hydroxyvitamin D(3) 1-alpha-hydroxylase) (VD3 1A hydroxylase) (P450C1 alpha) (P450VD1-alpha). | |||||
|
CP26B_HUMAN
|
||||||
| θ value | 1.74391e-19 (rank : 42) | NC score | 0.864171 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9NR63, Q53TW1, Q9NP41 | Gene names | CYP26B1, CYP26A2, P450RAI2 | |||
|
Domain Architecture |
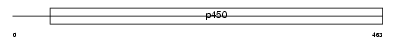
|
|||||
| Description | Cytochrome P450 26B1 (EC 1.14.-.-) (P450 26A2) (P450 retinoic acid- inactivating 2) (P450RAI-2) (Retinoic acid-metabolizing cytochrome). | |||||
|
CP27A_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 43) | NC score | 0.866594 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q02318, Q6LDB4, Q86YQ6 | Gene names | CYP27A1, CYP27 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 27, mitochondrial precursor (EC 1.14.13.15) (Cytochrome P-450C27/25) (Sterol 26-hydroxylase) (Sterol 27- hydroxylase) (Vitamin D(3) 25-hydroxylase) (5-beta-cholestane-3- alpha,7-alpha,12-alpha-triol 27-hydroxylase). | |||||
|
CP27A_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 44) | NC score | 0.868761 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9DBG1 | Gene names | Cyp27a1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 27, mitochondrial precursor (EC 1.14.13.15) (Cytochrome P-450C27/25) (Sterol 26-hydroxylase) (Sterol 27- hydroxylase) (Vitamin D(3) 25-hydroxylase) (5-beta-cholestane-3- alpha,7-alpha,12-alpha-triol 27-hydroxylase). | |||||
|
CP51A_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 45) | NC score | 0.899382 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q16850, O00770, O00772, Q16784, Q8N1A8, Q99868 | Gene names | CYP51A1, CYP51 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 51A1 (EC 1.14.13.70) (CYPLI) (P450LI) (Sterol 14-alpha demethylase) (Lanosterol 14-alpha demethylase) (LDM) (P450-14DM) (P45014DM). | |||||
|
CP2D9_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 46) | NC score | 0.773758 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P11714, Q64489 | Gene names | Cyp2d9, Cyp2d-9 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2D9 (EC 1.14.14.1) (CYPIID9) (P450-16-alpha) (CA) (Testosterone 16-alpha hydroxylase). | |||||
|
CP2B9_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 47) | NC score | 0.750824 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P12790 | Gene names | Cyp2b9, Cyp2b-9, Rip | |||
|
Domain Architecture |
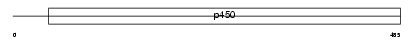
|
|||||
| Description | Cytochrome P450 2B9 (EC 1.14.14.1) (CYPIIB9) (Testosterone 16-alpha hydroxylase) (P450-16-alpha) (Clone PF26). | |||||
|
CP2B6_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 48) | NC score | 0.750470 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P20813, Q2V565, Q9UK46 | Gene names | CYP2B6 | |||
|
Domain Architecture |
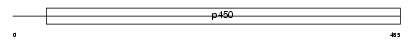
|
|||||
| Description | Cytochrome P450 2B6 (EC 1.14.14.1) (CYPIIB6) (P450 IIB1). | |||||
|
CP26A_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 49) | NC score | 0.873904 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O43174 | Gene names | CYP26A1, CYP26, P450RAI1 | |||
|
Domain Architecture |
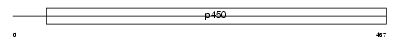
|
|||||
| Description | Cytochrome P450 26A1 (EC 1.14.-.-) (Retinoic acid-metabolizing cytochrome) (P450 retinoic acid-inactivating 1) (P450RAI) (hP450RAI) (Retinoic acid 4-hydroxylase). | |||||
|
CP2DQ_MOUSE
|
||||||
| θ value | 5.60996e-18 (rank : 50) | NC score | 0.760971 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q8CIM7, Q9DBJ5 | Gene names | Cyp2d26, Cyp2d-26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2D26 (EC 1.14.14.1) (CYPIID26). | |||||
|
CP2S1_MOUSE
|
||||||
| θ value | 7.32683e-18 (rank : 51) | NC score | 0.767668 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q9DBX6 | Gene names | Cyp2s1 | |||
|
Domain Architecture |
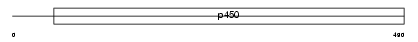
|
|||||
| Description | Cytochrome P450 2S1 (EC 1.14.14.1) (CYPIIS1). | |||||
|
CP2DA_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 52) | NC score | 0.768138 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P24456, Q64490 | Gene names | Cyp2d10, Cyp2d-10 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2D10 (EC 1.14.14.1) (CYPIID10) (P450-16-alpha) (P450CB) (Testosterone 16-alpha hydroxylase). | |||||
|
CP2D6_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 53) | NC score | 0.772856 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P10635, Q16752, Q6B012 | Gene names | CYP2D6 | |||
|
Domain Architecture |
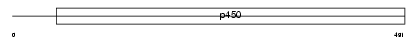
|
|||||
| Description | Cytochrome P450 2D6 (EC 1.14.14.1) (CYPIID6) (P450-DB1) (Debrisoquine 4-hydroxylase). | |||||
|
CP2BJ_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 54) | NC score | 0.756587 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | O55071 | Gene names | Cyp2b19 | |||
|
Domain Architecture |
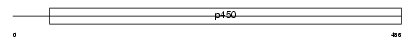
|
|||||
| Description | Cytochrome P450 2B19 (EC 1.14.14.1) (CYPIIB19). | |||||
|
CP2S1_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 55) | NC score | 0.763822 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q96SQ9, Q9BZ66 | Gene names | CYP2S1 | |||
|
Domain Architecture |
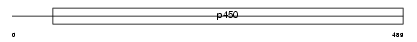
|
|||||
| Description | Cytochrome P450 2S1 (EC 1.14.14.1) (CYPIIS1). | |||||
|
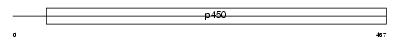
CP26A_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 56) | NC score | 0.875980 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O55127, Q9R1F4 | Gene names | Cyp26a1, Cyp26, P450ra | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 26A1 (EC 1.14.-.-) (Retinoic acid-metabolizing cytochrome) (P450RAI) (Retinoic acid 4-hydroxylase). | |||||
|
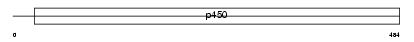
CP239_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 57) | NC score | 0.753141 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P56656 | Gene names | Cyp2c39 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 2C39 (EC 1.14.14.1) (CYPIIC39). | |||||
|
CP2W1_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 58) | NC score | 0.776456 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q8TAV3 | Gene names | CYP2W1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2W1 (EC 1.14.14.-) (CYPIIW1). | |||||
|
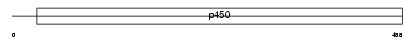
CP2A4_MOUSE
|
||||||
| θ value | 1.38178e-16 (rank : 59) | NC score | 0.742732 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P15392 | Gene names | Cyp2a4, Cyp2a-4 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 2A4 (EC 1.14.14.1) (CYPIIA4) (Testosterone 15-alpha- hydroxylase) (P450-15-alpha) (P450-IIA3.1). | |||||
|
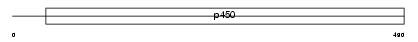
CP17A_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 60) | NC score | 0.829430 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P05093, Q5TZV7 | Gene names | CYP17A1, CYP17, S17AH | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 17A1 (EC 1.14.99.9) (CYPXVII) (P450-C17) (P450c17) (Steroid 17-alpha-monooxygenase) (Steroid 17-alpha-hydroxylase/17,20 lyase). | |||||
|
CP2A6_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 61) | NC score | 0.749046 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P11509, P00190, P10890, Q16803, Q4VAT9, Q4VAU0, Q4VAU1, Q9H1Z7, Q9UK48 | Gene names | CYP2A6 | |||
|
Domain Architecture |
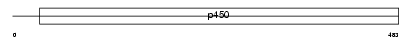
|
|||||
| Description | Cytochrome P450 2A6 (EC 1.14.14.1) (CYPIIA6) (Coumarin 7-hydroxylase) (P450 IIA3) (CYP2A3) (P450(I)). | |||||
|
CP2DB_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 62) | NC score | 0.750057 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P24457 | Gene names | Cyp2d11, Cyp2d-11 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2D11 (EC 1.14.14.1) (CYPIID11) (P450-16-alpha) (CC) (Testosterone 16-alpha hydroxylase). | |||||
|
CP2A7_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 63) | NC score | 0.743235 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P20853, Q13121 | Gene names | CYP2A7 | |||
|
Domain Architecture |
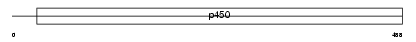
|
|||||
| Description | Cytochrome P450 2A7 (EC 1.14.14.1) (CYPIIA7) (P450-IIA4). | |||||
|
CP2F2_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 64) | NC score | 0.749542 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P33267 | Gene names | Cyp2f2, Cyp2f-2 | |||
|
Domain Architecture |
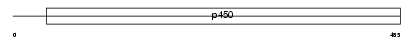
|
|||||
| Description | Cytochrome P450 2F2 (EC 1.14.14.-) (CYPIIF2) (Naphthalene dehydrogenase) (Naphthalene hydroxylase) (P450-NAH-2). | |||||
|
CP2A5_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 65) | NC score | 0.744827 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P20852 | Gene names | Cyp2a5, Cyp2a-5 | |||
|
Domain Architecture |
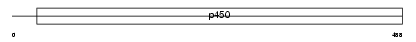
|
|||||
| Description | Cytochrome P450 2A5 (EC 1.14.14.1) (CYPIIA5) (Coumarin 7-hydroxylase) (P450-15-COH) (P450-IIA3.2). | |||||
|
CP2AD_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 66) | NC score | 0.738037 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q16696, Q53YR8, Q9H2X2 | Gene names | CYP2A13 | |||
|
Domain Architecture |
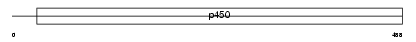
|
|||||
| Description | Cytochrome P450 2A13 (EC 1.14.14.1) (CYPIIA13). | |||||
|
CP2AC_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 67) | NC score | 0.739590 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P56593 | Gene names | Cyp2a12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2A12 (EC 1.14.14.1) (CYPIIA12) (Steroid hormones 7- alpha-hydroxylase) (Testosterone 7-alpha-hydroxylase). | |||||
|
CP2CT_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 68) | NC score | 0.739750 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q64458, Q8WUN8 | Gene names | Cyp2c29 | |||
|
Domain Architecture |
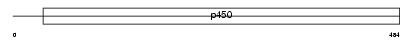
|
|||||
| Description | Cytochrome P450 2C29 (EC 1.14.14.1) (CYPIIC29) (P-450 MUT-2) (Aldehyde oxygenase). | |||||
|
CP21A_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 69) | NC score | 0.824558 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P03940, O88304, Q64510, Q9QX14 | Gene names | Cyp21, Cyp21a-1, Cyp21a1 | |||
|
Domain Architecture |
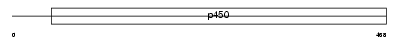
|
|||||
| Description | Cytochrome P450 21 (EC 1.14.99.10) (Cytochrome P450 XXI) (Steroid 21- hydroxylase) (21-OHase) (P450-C21) (P-450c21). | |||||
|
CP2BA_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 70) | NC score | 0.739167 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | P12791 | Gene names | Cyp2b10, Cyp2b-10 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2B10 (EC 1.14.14.1) (CYPIIB10) (Testosterone 16-alpha hydroxylase) (P450-16-alpha) (Clone PF3/46). | |||||
|
CP21A_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 71) | NC score | 0.834566 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P08686, P04033, Q01204, Q16749 | Gene names | CYP21A2, CYP21, CYP21B | |||
|
Domain Architecture |
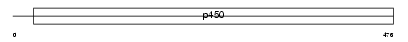
|
|||||
| Description | Cytochrome P450 21 (EC 1.14.99.10) (Cytochrome P450 XXI) (Steroid 21- hydroxylase) (21-OHase) (P450-C21) (P-450c21) (P450-C21B). | |||||
|
CP1A2_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 72) | NC score | 0.785231 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P05177, Q16754, Q6NWU5, Q9BXX7, Q9UK49 | Gene names | CYP1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1A2 (EC 1.14.14.1) (CYPIA2) (P450-P3) (P(3)450) (P450 4). | |||||
|
CP1A2_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 73) | NC score | 0.768276 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P00186, Q9QWJ4 | Gene names | Cyp1a2, Cyp1a-2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1A2 (EC 1.14.14.1) (CYPIA2) (P450-P2/P450-P3). | |||||
|
CP2CI_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 74) | NC score | 0.742894 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P33260, Q16703, Q16751 | Gene names | CYP2C18 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 2C18 (EC 1.14.14.1) (CYPIIC18) (P450-6B/29C). | |||||
|
CP2F1_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 75) | NC score | 0.744662 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P24903 | Gene names | CYP2F1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 2F1 (EC 1.14.14.1) (CYPIIF1). | |||||
|
CP19A_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 76) | NC score | 0.853315 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P28649 | Gene names | Cyp19a1, Arom, Cyp19 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 19A1 (EC 1.14.14.1) (Aromatase) (CYPXIX) (Estrogen synthetase) (P-450AROM). | |||||
|
CP1B1_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 77) | NC score | 0.771697 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q64429, Q60593, Q64461 | Gene names | Cyp1b1, Cyp1-b1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 1B1 (EC 1.14.14.1) (CYPIB1) (P450CMEF) (P450EF). | |||||
|
CP2C9_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 78) | NC score | 0.753703 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P11712, P11713, Q16756, Q16872 | Gene names | CYP2C9, CYP2C10 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C9 (EC 1.14.13.80) ((R)-limonene 6-monooxygenase) (EC 1.14.13.48) ((S)-limonene 6-monooxygenase) (EC 1.14.13.49) ((S)- limonene 7-monooxygenase) (CYPIIC9) (P450 PB-1) (P450 MP-4/MP-8) (S- mephenytoin 4-hydroxylase) (P-450MP). | |||||
|
CP11A_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 79) | NC score | 0.829050 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9QZ82 | Gene names | Cyp11a1, Cyp11a | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11A1, mitochondrial precursor (EC 1.14.15.6) (CYPXIA1) (P450(scc)) (Cholesterol side-chain cleavage enzyme) (Cholesterol desmolase). | |||||
|
CP1A1_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 80) | NC score | 0.785642 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P00184, Q61455 | Gene names | Cyp1a1, Cyp1a-1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1A1 (EC 1.14.14.1) (CYPIA1) (P450-P1). | |||||
|
CP238_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 81) | NC score | 0.737238 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P56655 | Gene names | Cyp2c38 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 2C38 (EC 1.14.14.1) (CYPIIC38). | |||||
|
CP270_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 82) | NC score | 0.756268 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q91W64, Q5GLY9, Q80VW0, Q8R120, Q8VC00 | Gene names | Cyp2c70, Cyp2c-70, Cyp2c22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2C70 (EC 1.14.14.1) (CYPIIC70). | |||||
|
CP2CJ_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 83) | NC score | 0.749226 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P33261, P33259, Q8WZB1, Q8WZB2 | Gene names | CYP2C19 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 2C19 (EC 1.14.13.80) ((R)-limonene 6-monooxygenase) (EC 1.14.13.48) ((S)-limonene 6-monooxygenase) (EC 1.14.13.49) ((S)- limonene 7-monooxygenase) (CYPIIC19) (P450-11A) (Mephenytoin 4- hydroxylase) (CYPIIC17) (P450-254C). | |||||
|
CP2E1_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 84) | NC score | 0.748657 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q05421, Q9Z198 | Gene names | Cyp2e1, Cyp2e, Cyp2e-1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 2E1 (EC 1.14.14.1) (CYPIIE1) (P450-J) (P450-ALC). | |||||
|
CP11A_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 85) | NC score | 0.817572 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P05108, Q15081, Q16805 | Gene names | CYP11A1, CYP11A | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 11A1, mitochondrial precursor (EC 1.14.15.6) (CYPXIA1) (P450(scc)) (Cholesterol side-chain cleavage enzyme) (Cholesterol desmolase). | |||||
|
CP2E1_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 86) | NC score | 0.752131 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P05181, Q9UK47 | Gene names | CYP2E1, CYP2E | |||
|
Domain Architecture |
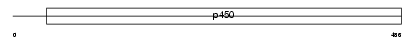
|
|||||
| Description | Cytochrome P450 2E1 (EC 1.14.14.1) (CYPIIE1) (P450-J). | |||||
|
CP19A_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 87) | NC score | 0.871473 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P11511, Q16731, Q58FA0 | Gene names | CYP19A1, ARO1, CYAR, CYP19 | |||
|
Domain Architecture |
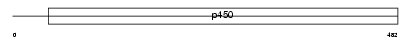
|
|||||
| Description | Cytochrome P450 19A1 (EC 1.14.14.1) (Aromatase) (CYPXIX) (Estrogen synthetase) (P-450AROM). | |||||
|
CP1A1_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 88) | NC score | 0.772516 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P04798 | Gene names | CYP1A1 | |||
|
Domain Architecture |
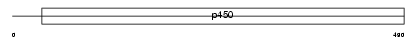
|
|||||
| Description | Cytochrome P450 1A1 (EC 1.14.14.1) (CYPIA1) (P450-P1) (P450 form 6) (P450-C). | |||||
|
CP2C8_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 89) | NC score | 0.744862 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P10632 | Gene names | CYP2C8 | |||
|
Domain Architecture |
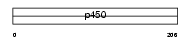
|
|||||
| Description | Cytochrome P450 2C8 (EC 1.14.14.1) (CYPIIC8) (P450 form 1) (P450 MP- 12/MP-20) (P450 IIC2) (S-mephenytoin 4-hydroxylase). | |||||
|
CP1B1_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 90) | NC score | 0.765314 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q16678, Q93089, Q9H316 | Gene names | CYP1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1B1 (EC 1.14.14.1) (CYPIB1). | |||||
|
CP237_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 91) | NC score | 0.738399 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P56654 | Gene names | Cyp2c37 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 2C37 (EC 1.14.14.1) (CYPIIC37). | |||||
|
CP39A_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 92) | NC score | 0.774775 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q9NYL5, Q96FW5 | Gene names | CYP39A1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (hCYP39A1). | |||||
|
CP240_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 93) | NC score | 0.739698 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P56657 | Gene names | Cyp2c40 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 2C40 (EC 1.14.14.1) (CYPIIC40). | |||||
|
CP39A_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 94) | NC score | 0.767306 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q9JKJ9 | Gene names | Cyp39a1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (mCYP39A1). | |||||
|
C11B1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 95) | NC score | 0.804928 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P15538, Q14095, Q9UML2 | Gene names | CYP11B1, S11BH | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11B1, mitochondrial precursor (EC 1.14.15.4) (CYPXIB1) (P450C11) (P-450c11) (Steroid 11-beta-hydroxylase). | |||||
|
CP26C_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 96) | NC score | 0.823535 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q6V0L0, Q5VXH6 | Gene names | CYP26C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 26C1 (EC 1.14.-.-). | |||||
|
C11B2_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 97) | NC score | 0.805615 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P19099, Q16726 | Gene names | CYP11B2 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 11B2, mitochondrial precursor (EC 1.14.15.4) (EC 1.14.15.5) (CYPXIB2) (P-450Aldo) (Aldosterone synthase) (ALDOS) (Aldosterone-synthesizing enzyme) (Steroid 18-hydroxylase) (P-450C18). | |||||
|
C11B2_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 98) | NC score | 0.786212 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P15539, Q64661 | Gene names | Cyp11b2, Cyp11b-2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11B2, mitochondrial precursor (EC 1.14.15.4) (EC 1.14.15.5) (CYPXIB2) (P450C11) (Steroid 11-beta-hydroxylase) (Aldosterone synthase). | |||||
|
CP7A1_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 99) | NC score | 0.625202 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P22680, P78454, Q7KZ19 | Gene names | CYP7A1, CYP7 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 7A1 (Cholesterol 7-alpha-monooxygenase) (CYPVII) (EC 1.14.13.17) (Cholesterol 7-alpha-hydroxylase). | |||||
|
CP7A1_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 100) | NC score | 0.621004 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q64505 | Gene names | Cyp7a1, Cyp7 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 7A1 (EC 1.14.13.17) (Cholesterol 7-alpha- monooxygenase) (CYPVII) (Cholesterol 7-alpha-hydroxylase). | |||||
|
CP7B1_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 101) | NC score | 0.647580 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | O75881, Q9UNF5 | Gene names | CYP7B1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 7B1 (EC 1.14.13.100) (25-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase). | |||||
|
CP7B1_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 102) | NC score | 0.644676 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q60991 | Gene names | Cyp7b1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 7B1 (EC 1.14.13.100) (25-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (HCT-1). | |||||
|
CP8B1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 103) | NC score | 0.577925 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9UNU6, O75958 | Gene names | CYP8B1, CYP12 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 8B1 (EC 1.14.13.95) (CYPVIIIB1) (7-alpha- hydroxycholest-4-en-3-one 12-alpha-hydroxylase) (Sterol 12-alpha- hydroxylase) (7-alpha-hydroxy-4-cholesten-3-one 12-alpha-hydroxylase). | |||||
|
PTGIS_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 104) | NC score | 0.574171 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | O35074 | Gene names | Ptgis, Cyp8 | |||
|
Domain Architecture |
|
|||||
| Description | Prostacyclin synthase (EC 5.3.99.4) (Prostaglandin I2 synthase). | |||||
|
CP8B1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 105) | NC score | 0.550576 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O88962, Q9R217 | Gene names | Cyp8b1, Cyp12 | |||
|
Domain Architecture |
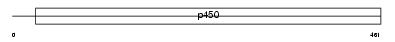
|
|||||
| Description | Cytochrome P450 8B1 (EC 1.14.13.95) (CYPVIIIB1) (7-alpha- hydroxycholest-4-en-3-one 12-alpha-hydroxylase) (Sterol 12-alpha- hydroxylase) (7-alpha-hydroxy-4-cholesten-3-one 12-alpha-hydroxylase). | |||||
|
CENPK_HUMAN
|
||||||
| θ value | 0.125558 (rank : 106) | NC score | 0.015122 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BS16, Q9H4L0 | Gene names | CENPK, ICEN37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein K (CENP-K) (Interphase centromere complex protein 37) (Protein AF-5alpha) (p33). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 107) | NC score | -0.001890 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 108) | NC score | -0.001105 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
STK10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | -0.011793 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
UBE4B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.009146 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
UBE4B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.010246 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ES00, Q9EQE0 | Gene names | Ube4b, Ufd2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Ufd2a). | |||||
|
AXUD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.004637 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59054 | Gene names | Axud1, Taip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axin-1 up-regulated gene 1 protein (TGF-beta-induced apoptosis protein 3) (TAIP-3). | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | -0.001491 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
COX3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.006578 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00414 | Gene names | MT-CO3, COIII, COXIII, MTCO3 | |||
|
Domain Architecture |
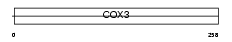
|
|||||
| Description | Cytochrome c oxidase subunit 3 (EC 1.9.3.1) (Cytochrome c oxidase polypeptide III). | |||||
|
SPAT7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.005370 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P0W8, Q8WX30, Q96HF3, Q9H0X0, Q9P0W7 | Gene names | SPATA7, HSD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 7 (Spermatogenesis-associated protein HSD3) (HSD-3.1). | |||||
|
DHC24_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.006416 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15392, Q9HBA8 | Gene names | DHCR24, KIAA0018 | |||
|
Domain Architecture |
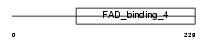
|
|||||
| Description | 24-dehydrocholesterol reductase precursor (EC 1.3.1.-) (3-beta- hydroxysterol delta-24-reductase) (Seladin-1) (Diminuto/dwarf1 homolog). | |||||
|
UB2Q2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.002917 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2Z8, Q3UBX3, Q3V3A5, Q8BUN2, Q8BVX5 | Gene names | Ube2q2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 Q2 (EC 6.3.2.19) (Ubiquitin-protein ligase Q2) (Ubiquitin carrier protein Q2). | |||||
|
PTGIS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.508017 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q16647, Q9HAX2, Q9HAX3, Q9HAX4 | Gene names | PTGIS, CYP8, CYP8A1 | |||
|
Domain Architecture |
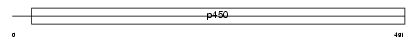
|
|||||
| Description | Prostacyclin synthase (EC 5.3.99.4) (Prostaglandin I2 synthase). | |||||
|
CP4Z1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q86W10 | Gene names | CYP4Z1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4Z1 (EC 1.14.14.1) (CYPIVZ1). | |||||
|
CP4X1_HUMAN
|
||||||
| NC score | 0.995597 (rank : 2) | θ value | 1.91371e-159 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q8N118 | Gene names | CYP4X1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4X1 (EC 1.14.14.1) (CYPIVX1). | |||||
|
CP4AE_MOUSE
|
||||||
| NC score | 0.993840 (rank : 3) | θ value | 1.98959e-140 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O35728, Q3UER2, Q5EBH0 | Gene names | Cyp4a14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4A14 precursor (EC 1.14.15.3) (CYPIVA14) (Lauric acid omega-hydroxylase 3) (P450-LA-omega 3). | |||||
|
CP4AA_MOUSE
|
||||||
| NC score | 0.993311 (rank : 4) | θ value | 7.05993e-146 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O88833, Q9DCS5 | Gene names | Cyp4a10, Cyp4a-10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4A10 (EC 1.14.15.3) (CYPIVA10) (Lauric acid omega- hydroxylase) (P450-LA-omega 1) (P452). | |||||
|
CP4AB_HUMAN
|
||||||
| NC score | 0.993024 (rank : 5) | θ value | 8.33966e-147 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q02928, Q06766, Q16865, Q16866, Q8IWY5 | Gene names | CYP4A11 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4A11 precursor (EC 1.14.15.3) (CYPIVA11) (Fatty acid omega-hydroxylase) (P-450 HK omega) (Lauric acid omega-hydroxylase) (CYP4AII) (P450-HL-omega). | |||||
|
CP4AC_MOUSE
|
||||||
| NC score | 0.992635 (rank : 6) | θ value | 5.05951e-144 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q91WL5, Q3UNE4, Q6P931, Q8N7N3 | Gene names | Cyp4a12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4A12 (EC 1.14.14.1) (CYPIVA12). | |||||
|
CP4B1_MOUSE
|
||||||
| NC score | 0.990630 (rank : 7) | θ value | 4.74654e-134 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q64462 | Gene names | Cyp4b1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4B1 (EC 1.14.14.1) (CYPIVB1). | |||||
|
CP4B1_HUMAN
|
||||||
| NC score | 0.990581 (rank : 8) | θ value | 1.47893e-127 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P13584, Q8TD85, Q8WWU9, Q8WWV0 | Gene names | CYP4B1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4B1 (EC 1.14.14.1) (CYPIVB1) (P450-HP). | |||||
|
CP4FC_HUMAN
|
||||||
| NC score | 0.985092 (rank : 9) | θ value | 4.33309e-103 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9HCS2, Q9HCS1 | Gene names | CYP4F12 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4F12 (EC 1.14.14.1) (CYPIVF12). | |||||
|
CP4F8_HUMAN
|
||||||
| NC score | 0.985073 (rank : 10) | θ value | 6.68496e-104 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P98187 | Gene names | CYP4F8 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4F8 (EC 1.14.14.1) (CYPIVF8). | |||||
|
CP4FB_HUMAN
|
||||||
| NC score | 0.984599 (rank : 11) | θ value | 1.48926e-103 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9HBI6, O75254, Q96AQ5 | Gene names | CYP4F11 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4F11 (EC 1.14.14.1) (CYPIVF11). | |||||
|
CP4F2_HUMAN
|
||||||
| NC score | 0.984303 (rank : 12) | θ value | 7.89671e-105 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P78329, Q16677, Q6NWT4, Q6NWT6, Q9NNZ0, Q9UIU8 | Gene names | CYP4F2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4F2 (EC 1.14.13.30) (CYPIVF2) (Leukotriene-B(4) omega- hydroxylase) (Leukotriene-B(4) 20-monooxygenase) (Cytochrome P450-LTB- omega). | |||||
|
CP4F3_HUMAN
|
||||||
| NC score | 0.983776 (rank : 13) | θ value | 3.4333e-100 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q08477, O60634, Q5U740 | Gene names | CYP4F3, LTB4H | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4F3 (EC 1.14.13.30) (CYPIVF3) (Leukotriene-B(4) omega- hydroxylase) (Leukotriene-B(4) 20-monooxygenase) (Cytochrome P450-LTB- omega). | |||||
|
CP4FE_MOUSE
|
||||||
| NC score | 0.982860 (rank : 14) | θ value | 4.33309e-103 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9EP75, Q52L93 | Gene names | Cyp4f14 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4F14 (EC 1.14.13.30) (Leukotriene-B4 omega- hydroxylase) (Leukotriene-B4 20-monooxygenase) (Cytochrome P450-LTB- omega) (Cyp4f-14). | |||||
|
CP4F3_MOUSE
|
||||||
| NC score | 0.981424 (rank : 15) | θ value | 6.47494e-99 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q99N16, Q9D8N4 | Gene names | Cyp4f3, Cyp4f18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4F3 (EC 1.14.13.30) (CYPIVF3) (Leukotriene-B(4) omega- hydroxylase) (Leukotriene-B(4) 20-monooxygenase) (Cytochrome P450-LTB- omega). | |||||
|
CP4V2_HUMAN
|
||||||
| NC score | 0.968523 (rank : 16) | θ value | 1.31987e-59 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q6ZWL3, Q6ZTM4 | Gene names | CYP4V2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4V2 (EC 1.14.-.-). | |||||
|
CP4V3_MOUSE
|
||||||
| NC score | 0.968322 (rank : 17) | θ value | 1.84173e-61 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9DBW0 | Gene names | Cyp4v3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4V3 (EC 1.14.-.-). | |||||
|
CP46A_MOUSE
|
||||||
| NC score | 0.924896 (rank : 18) | θ value | 8.63488e-27 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9WVK8 | Gene names | Cyp46a1, Cyp46 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome p450 46A1 (EC 1.14.13.98) (Cholesterol 24-hydroxylase) (CH24H). | |||||
|
CP46A_HUMAN
|
||||||
| NC score | 0.923007 (rank : 19) | θ value | 5.59698e-26 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9Y6A2 | Gene names | CYP46A1, CYP46 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome p450 46A1 (EC 1.14.13.98) (Cholesterol 24-hydroxylase) (CH24H). | |||||
|
CP3AD_MOUSE
|
||||||
| NC score | 0.916699 (rank : 20) | θ value | 9.50281e-42 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q64464 | Gene names | Cyp3a13, Cyp3a-13 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 3A13 (EC 1.14.14.1) (CYPIIIA13). | |||||
|
CP3AP_MOUSE
|
||||||
| NC score | 0.916368 (rank : 21) | θ value | 3.26607e-42 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O09158 | Gene names | Cyp3a25 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 3A25 (EC 1.14.14.1) (CYPIIIA25). | |||||
|
CP343_HUMAN
|
||||||
| NC score | 0.915394 (rank : 22) | θ value | 1.01529e-43 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9HB55, Q9HB52, Q9HB53, Q9HB54, Q9HB57 | Gene names | CYP3A43 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 3A43 (EC 1.14.14.1). | |||||
|
THAS_MOUSE
|
||||||
| NC score | 0.915290 (rank : 23) | θ value | 4.89182e-30 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P36423, P97505 | Gene names | Tbxas1, Cyp5, Cyp5a1 | |||
|
Domain Architecture |

|
|||||
| Description | Thromboxane-A synthase (EC 5.3.99.5) (TXA synthase) (TXS) (TS) (Cytochrome P450 5A1). | |||||
|
CP3A7_HUMAN
|
||||||
| NC score | 0.912831 (rank : 24) | θ value | 1.91475e-42 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P24462 | Gene names | CYP3A7 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 3A7 (EC 1.14.14.1) (CYPIIIA7) (P450-HFLA). | |||||
|
CP3A4_HUMAN
|
||||||
| NC score | 0.911627 (rank : 25) | θ value | 2.11701e-41 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P08684, Q16757, Q9UK50 | Gene names | CYP3A4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 3A4 (EC 1.14.13.67) (Quinine 3-monooxygenase) (CYPIIIA4) (Nifedipine oxidase) (Taurochenodeoxycholate 6-alpha- hydroxylase) (EC 1.14.13.97) (NF-25) (P450-PCN1). | |||||
|
CP3AG_MOUSE
|
||||||
| NC score | 0.911589 (rank : 26) | θ value | 1.6774e-38 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q64481 | Gene names | Cyp3a16, Cyp3a-16 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 3A16 (EC 1.14.14.1) (CYPIIIA16). | |||||
|
CP341_MOUSE
|
||||||
| NC score | 0.910738 (rank : 27) | θ value | 8.89434e-40 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9JMA7 | Gene names | Cyp3a41 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 3A41 (EC 1.14.14.1). | |||||
|
CP3A5_HUMAN
|
||||||
| NC score | 0.909956 (rank : 28) | θ value | 3.73686e-38 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P20815, Q53WY8, Q75MV0, Q9HB56 | Gene names | CYP3A5 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 3A5 (EC 1.14.14.1) (CYPIIIA5) (P450-PCN3) (HLp2). | |||||
|
CP3A3_HUMAN
|
||||||
| NC score | 0.909872 (rank : 29) | θ value | 8.04462e-41 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P05184 | Gene names | CYP3A3 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 3A3 (EC 1.14.14.1) (CYPIIIA3) (HLp). | |||||
|
CP3AB_MOUSE
|
||||||
| NC score | 0.909548 (rank : 30) | θ value | 3.37985e-39 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q64459 | Gene names | Cyp3a11, Cyp3a-11 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 3A11 (EC 1.14.14.1) (CYPIIIA11) (P-450IIIAM1) (P- 450UT). | |||||
|
THAS_HUMAN
|
||||||
| NC score | 0.903184 (rank : 31) | θ value | 6.84181e-24 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P24557, Q16844, Q8IUN1, Q9GZW4, Q9HD77, Q9HD78, Q9HD79, Q9HD80, Q9HD81, Q9HD82, Q9HD83, Q9HD84 | Gene names | TBXAS1, CYP5, CYP5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Thromboxane-A synthase (EC 5.3.99.5) (TXA synthase) (TXS) (Cytochrome P450 5A1). | |||||
|
CP51A_HUMAN
|
||||||
| NC score | 0.899382 (rank : 32) | θ value | 1.13037e-18 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q16850, O00770, O00772, Q16784, Q8N1A8, Q99868 | Gene names | CYP51A1, CYP51 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 51A1 (EC 1.14.13.70) (CYPLI) (P450LI) (Sterol 14-alpha demethylase) (Lanosterol 14-alpha demethylase) (LDM) (P450-14DM) (P45014DM). | |||||
|
CP26A_MOUSE
|
||||||
| NC score | 0.875980 (rank : 33) | θ value | 6.20254e-17 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O55127, Q9R1F4 | Gene names | Cyp26a1, Cyp26, P450ra | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 26A1 (EC 1.14.-.-) (Retinoic acid-metabolizing cytochrome) (P450RAI) (Retinoic acid 4-hydroxylase). | |||||
|
CP26A_HUMAN
|
||||||
| NC score | 0.873904 (rank : 34) | θ value | 4.29542e-18 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O43174 | Gene names | CYP26A1, CYP26, P450RAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 26A1 (EC 1.14.-.-) (Retinoic acid-metabolizing cytochrome) (P450 retinoic acid-inactivating 1) (P450RAI) (hP450RAI) (Retinoic acid 4-hydroxylase). | |||||
|
CP24A_HUMAN
|
||||||
| NC score | 0.873365 (rank : 35) | θ value | 1.29031e-22 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q07973, Q15807 | Gene names | CYP24A1, CYP24 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 24A1, mitochondrial precursor (EC 1.14.-.-) (P450- CC24) (Vitamin D(3) 24-hydroxylase) (1,25-dihydroxyvitamin D(3) 24- hydroxylase) (24-OHase). | |||||
|
CP27B_HUMAN
|
||||||
| NC score | 0.872820 (rank : 36) | θ value | 4.15078e-21 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O15528 | Gene names | CYP27B1, CYP1ALPHA, CYP27B | |||
|
Domain Architecture |

|
|||||
| Description | 25-hydroxyvitamin D-1 alpha hydroxylase, mitochondrial precursor (EC 1.14.13.13) (Cytochrome P450 subfamily XXVIIB polypeptide 1) (Cytochrome p450 27B1) (Calcidiol 1-monooxygenase) (25-OHD-1 alpha- hydroxylase) (25-hydroxyvitamin D(3) 1-alpha-hydroxylase) (VD3 1A hydroxylase) (P450C1 alpha) (P450VD1-alpha). | |||||
|
CP27B_MOUSE
|
||||||
| NC score | 0.872750 (rank : 37) | θ value | 8.36355e-22 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O35084 | Gene names | Cyp27b1, Cyp27b, Cyp40 | |||
|
Domain Architecture |

|
|||||
| Description | 25-hydroxyvitamin D-1 alpha hydroxylase, mitochondrial precursor (EC 1.14.13.13) (Cytochrome P450 subfamily XXVIIB polypeptide 1) (Cytochrome p450 27B1) (Calcidiol 1-monooxygenase) (25-OHD-1 alpha- hydroxylase) (25-hydroxyvitamin D(3) 1-alpha-hydroxylase) (VD3 1A hydroxylase) (P450C1 alpha) (P450VD1-alpha). | |||||
|
CP24A_MOUSE
|
||||||
| NC score | 0.871528 (rank : 38) | θ value | 4.43474e-23 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q64441 | Gene names | Cyp24a1, Cyp-24, Cyp24 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 24A1, mitochondrial precursor (EC 1.14.-.-) (P450- CC24) (Vitamin D(3) 24-hydroxylase) (1,25-dihydroxyvitamin D(3) 24- hydroxylase) (24-OHase). | |||||
|
CP19A_HUMAN
|
||||||
| NC score | 0.871473 (rank : 39) | θ value | 3.18553e-13 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P11511, Q16731, Q58FA0 | Gene names | CYP19A1, ARO1, CYAR, CYP19 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 19A1 (EC 1.14.14.1) (Aromatase) (CYPXIX) (Estrogen synthetase) (P-450AROM). | |||||
|
CP27A_MOUSE
|
||||||
| NC score | 0.868761 (rank : 40) | θ value | 3.88503e-19 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9DBG1 | Gene names | Cyp27a1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 27, mitochondrial precursor (EC 1.14.13.15) (Cytochrome P-450C27/25) (Sterol 26-hydroxylase) (Sterol 27- hydroxylase) (Vitamin D(3) 25-hydroxylase) (5-beta-cholestane-3- alpha,7-alpha,12-alpha-triol 27-hydroxylase). | |||||
|
CP27A_HUMAN
|
||||||
| NC score | 0.866594 (rank : 41) | θ value | 2.27762e-19 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q02318, Q6LDB4, Q86YQ6 | Gene names | CYP27A1, CYP27 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 27, mitochondrial precursor (EC 1.14.13.15) (Cytochrome P-450C27/25) (Sterol 26-hydroxylase) (Sterol 27- hydroxylase) (Vitamin D(3) 25-hydroxylase) (5-beta-cholestane-3- alpha,7-alpha,12-alpha-triol 27-hydroxylase). | |||||
|
CP26B_HUMAN
|
||||||
| NC score | 0.864171 (rank : 42) | θ value | 1.74391e-19 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9NR63, Q53TW1, Q9NP41 | Gene names | CYP26B1, CYP26A2, P450RAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 26B1 (EC 1.14.-.-) (P450 26A2) (P450 retinoic acid- inactivating 2) (P450RAI-2) (Retinoic acid-metabolizing cytochrome). | |||||
|
CP19A_MOUSE
|
||||||
| NC score | 0.853315 (rank : 43) | θ value | 2.88119e-14 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P28649 | Gene names | Cyp19a1, Arom, Cyp19 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 19A1 (EC 1.14.14.1) (Aromatase) (CYPXIX) (Estrogen synthetase) (P-450AROM). | |||||
|
CP21A_HUMAN
|
||||||
| NC score | 0.834566 (rank : 44) | θ value | 3.40345e-15 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P08686, P04033, Q01204, Q16749 | Gene names | CYP21A2, CYP21, CYP21B | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 21 (EC 1.14.99.10) (Cytochrome P450 XXI) (Steroid 21- hydroxylase) (21-OHase) (P450-C21) (P-450c21) (P450-C21B). | |||||
|
CP17A_MOUSE
|
||||||
| NC score | 0.832550 (rank : 45) | θ value | 6.40375e-22 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P27786 | Gene names | Cyp17a1, Cyp17 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 17A1 (EC 1.14.99.9) (CYPXVII) (P450-C17) (P450c17) (Steroid 17-alpha-hydroxylase/17,20 lyase). | |||||
|
CP17A_HUMAN
|
||||||
| NC score | 0.829430 (rank : 46) | θ value | 1.80466e-16 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P05093, Q5TZV7 | Gene names | CYP17A1, CYP17, S17AH | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 17A1 (EC 1.14.99.9) (CYPXVII) (P450-C17) (P450c17) (Steroid 17-alpha-monooxygenase) (Steroid 17-alpha-hydroxylase/17,20 lyase). | |||||
|
CP11A_MOUSE
|
||||||
| NC score | 0.829050 (rank : 47) | θ value | 8.38298e-14 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9QZ82 | Gene names | Cyp11a1, Cyp11a | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11A1, mitochondrial precursor (EC 1.14.15.6) (CYPXIA1) (P450(scc)) (Cholesterol side-chain cleavage enzyme) (Cholesterol desmolase). | |||||
|
CP21A_MOUSE
|
||||||
| NC score | 0.824558 (rank : 48) | θ value | 1.99529e-15 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P03940, O88304, Q64510, Q9QX14 | Gene names | Cyp21, Cyp21a-1, Cyp21a1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 21 (EC 1.14.99.10) (Cytochrome P450 XXI) (Steroid 21- hydroxylase) (21-OHase) (P450-C21) (P-450c21). | |||||
|
CP26C_HUMAN
|
||||||
| NC score | 0.823535 (rank : 49) | θ value | 4.30538e-10 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q6V0L0, Q5VXH6 | Gene names | CYP26C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 26C1 (EC 1.14.-.-). | |||||
|
CP11A_HUMAN
|
||||||
| NC score | 0.817572 (rank : 50) | θ value | 2.43908e-13 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P05108, Q15081, Q16805 | Gene names | CYP11A1, CYP11A | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11A1, mitochondrial precursor (EC 1.14.15.6) (CYPXIA1) (P450(scc)) (Cholesterol side-chain cleavage enzyme) (Cholesterol desmolase). | |||||
|
CP2R1_HUMAN
|
||||||
| NC score | 0.815875 (rank : 51) | θ value | 1.05554e-24 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q6VVX0, Q5RT65 | Gene names | CYP2R1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2R1 (EC 1.14.14.-) (Vitamin D 25-hydroxylase). | |||||
|
CP2R1_MOUSE
|
||||||
| NC score | 0.811667 (rank : 52) | θ value | 8.36355e-22 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q6VVW9 | Gene names | Cyp2r1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2R1 (EC 1.14.14.-) (Vitamin D 25-hydroxylase). | |||||
|
C11B2_HUMAN
|
||||||
| NC score | 0.805615 (rank : 53) | θ value | 5.62301e-10 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P19099, Q16726 | Gene names | CYP11B2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11B2, mitochondrial precursor (EC 1.14.15.4) (EC 1.14.15.5) (CYPXIB2) (P-450Aldo) (Aldosterone synthase) (ALDOS) (Aldosterone-synthesizing enzyme) (Steroid 18-hydroxylase) (P-450C18). | |||||
|
C11B1_HUMAN
|
||||||
| NC score | 0.804928 (rank : 54) | θ value | 6.64225e-11 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P15538, Q14095, Q9UML2 | Gene names | CYP11B1, S11BH | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11B1, mitochondrial precursor (EC 1.14.15.4) (CYPXIB1) (P450C11) (P-450c11) (Steroid 11-beta-hydroxylase). | |||||
|
CP2J6_MOUSE
|
||||||
| NC score | 0.800522 (rank : 55) | θ value | 9.87957e-23 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O54750 | Gene names | Cyp2j6 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2J6 (EC 1.14.14.1) (CYPIIJ6) (Arachidonic acid epoxygenase). | |||||
|
CP2J5_MOUSE
|
||||||
| NC score | 0.800081 (rank : 56) | θ value | 4.9032e-22 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O54749 | Gene names | Cyp2j5 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2J5 (EC 1.14.14.1) (CYPIIJ5) (Arachidonic acid epoxygenase). | |||||
|
CP2J2_HUMAN
|
||||||
| NC score | 0.799806 (rank : 57) | θ value | 8.08199e-25 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P51589 | Gene names | CYP2J2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2J2 (EC 1.14.14.1) (CYPIIJ2) (Arachidonic acid epoxygenase). | |||||
|
C11B2_MOUSE
|
||||||
| NC score | 0.786212 (rank : 58) | θ value | 7.34386e-10 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P15539, Q64661 | Gene names | Cyp11b2, Cyp11b-2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11B2, mitochondrial precursor (EC 1.14.15.4) (EC 1.14.15.5) (CYPXIB2) (P450C11) (Steroid 11-beta-hydroxylase) (Aldosterone synthase). | |||||
|
CP1A1_MOUSE
|
||||||
| NC score | 0.785642 (rank : 59) | θ value | 8.38298e-14 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P00184, Q61455 | Gene names | Cyp1a1, Cyp1a-1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1A1 (EC 1.14.14.1) (CYPIA1) (P450-P1). | |||||
|
CP1A2_HUMAN
|
||||||
| NC score | 0.785231 (rank : 60) | θ value | 4.44505e-15 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P05177, Q16754, Q6NWU5, Q9BXX7, Q9UK49 | Gene names | CYP1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1A2 (EC 1.14.14.1) (CYPIA2) (P450-P3) (P(3)450) (P450 4). | |||||
|
CP2W1_HUMAN
|
||||||
| NC score | 0.776456 (rank : 61) | θ value | 1.058e-16 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q8TAV3 | Gene names | CYP2W1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2W1 (EC 1.14.14.-) (CYPIIW1). | |||||
|
CP39A_HUMAN
|
||||||
| NC score | 0.774775 (rank : 62) | θ value | 4.59992e-12 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q9NYL5, Q96FW5 | Gene names | CYP39A1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (hCYP39A1). | |||||
|
CP2D9_MOUSE
|
||||||
| NC score | 0.773758 (rank : 63) | θ value | 1.47631e-18 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P11714, Q64489 | Gene names | Cyp2d9, Cyp2d-9 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2D9 (EC 1.14.14.1) (CYPIID9) (P450-16-alpha) (CA) (Testosterone 16-alpha hydroxylase). | |||||
|
CP2D6_HUMAN
|
||||||
| NC score | 0.772856 (rank : 64) | θ value | 3.63628e-17 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P10635, Q16752, Q6B012 | Gene names | CYP2D6 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2D6 (EC 1.14.14.1) (CYPIID6) (P450-DB1) (Debrisoquine 4-hydroxylase). | |||||
|
CP1A1_HUMAN
|
||||||
| NC score | 0.772516 (rank : 65) | θ value | 3.18553e-13 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P04798 | Gene names | CYP1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1A1 (EC 1.14.14.1) (CYPIA1) (P450-P1) (P450 form 6) (P450-C). | |||||
|
CP1B1_MOUSE
|
||||||
| NC score | 0.771697 (rank : 66) | θ value | 3.76295e-14 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q64429, Q60593, Q64461 | Gene names | Cyp1b1, Cyp1-b1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1B1 (EC 1.14.14.1) (CYPIB1) (P450CMEF) (P450EF). | |||||
|
CP1A2_MOUSE
|
||||||
| NC score | 0.768276 (rank : 67) | θ value | 5.8054e-15 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P00186, Q9QWJ4 | Gene names | Cyp1a2, Cyp1a-2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1A2 (EC 1.14.14.1) (CYPIA2) (P450-P2/P450-P3). | |||||
|
CP2DA_MOUSE
|
||||||
| NC score | 0.768138 (rank : 68) | θ value | 2.13179e-17 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P24456, Q64490 | Gene names | Cyp2d10, Cyp2d-10 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2D10 (EC 1.14.14.1) (CYPIID10) (P450-16-alpha) (P450CB) (Testosterone 16-alpha hydroxylase). | |||||
|
CP2S1_MOUSE
|
||||||
| NC score | 0.767668 (rank : 69) | θ value | 7.32683e-18 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q9DBX6 | Gene names | Cyp2s1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2S1 (EC 1.14.14.1) (CYPIIS1). | |||||
|
CP39A_MOUSE
|
||||||
| NC score | 0.767306 (rank : 70) | θ value | 1.74796e-11 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q9JKJ9 | Gene names | Cyp39a1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (mCYP39A1). | |||||
|
CP1B1_HUMAN
|
||||||
| NC score | 0.765314 (rank : 71) | θ value | 9.26847e-13 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q16678, Q93089, Q9H316 | Gene names | CYP1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1B1 (EC 1.14.14.1) (CYPIB1). | |||||
|
CP2S1_HUMAN
|
||||||
| NC score | 0.763822 (rank : 72) | θ value | 4.74913e-17 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q96SQ9, Q9BZ66 | Gene names | CYP2S1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2S1 (EC 1.14.14.1) (CYPIIS1). | |||||
|
CP2DQ_MOUSE
|
||||||
| NC score | 0.760971 (rank : 73) | θ value | 5.60996e-18 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q8CIM7, Q9DBJ5 | Gene names | Cyp2d26, Cyp2d-26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2D26 (EC 1.14.14.1) (CYPIID26). | |||||
|
CP2BJ_MOUSE
|
||||||
| NC score | 0.756587 (rank : 74) | θ value | 4.74913e-17 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | O55071 | Gene names | Cyp2b19 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2B19 (EC 1.14.14.1) (CYPIIB19). | |||||
|
CP270_MOUSE
|
||||||
| NC score | 0.756268 (rank : 75) | θ value | 8.38298e-14 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q91W64, Q5GLY9, Q80VW0, Q8R120, Q8VC00 | Gene names | Cyp2c70, Cyp2c-70, Cyp2c22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2C70 (EC 1.14.14.1) (CYPIIC70). | |||||
|
CP2C9_HUMAN
|
||||||
| NC score | 0.753703 (rank : 76) | θ value | 4.91457e-14 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P11712, P11713, Q16756, Q16872 | Gene names | CYP2C9, CYP2C10 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C9 (EC 1.14.13.80) ((R)-limonene 6-monooxygenase) (EC 1.14.13.48) ((S)-limonene 6-monooxygenase) (EC 1.14.13.49) ((S)- limonene 7-monooxygenase) (CYPIIC9) (P450 PB-1) (P450 MP-4/MP-8) (S- mephenytoin 4-hydroxylase) (P-450MP). | |||||
|
CP239_MOUSE
|
||||||
| NC score | 0.753141 (rank : 77) | θ value | 8.10077e-17 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P56656 | Gene names | Cyp2c39 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C39 (EC 1.14.14.1) (CYPIIC39). | |||||
|
CP2E1_HUMAN
|
||||||
| NC score | 0.752131 (rank : 78) | θ value | 2.43908e-13 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P05181, Q9UK47 | Gene names | CYP2E1, CYP2E | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2E1 (EC 1.14.14.1) (CYPIIE1) (P450-J). | |||||
|
CP2B9_MOUSE
|
||||||
| NC score | 0.750824 (rank : 79) | θ value | 1.92812e-18 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P12790 | Gene names | Cyp2b9, Cyp2b-9, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2B9 (EC 1.14.14.1) (CYPIIB9) (Testosterone 16-alpha hydroxylase) (P450-16-alpha) (Clone PF26). | |||||
|
CP2B6_HUMAN
|
||||||
| NC score | 0.750470 (rank : 80) | θ value | 3.28887e-18 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P20813, Q2V565, Q9UK46 | Gene names | CYP2B6 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2B6 (EC 1.14.14.1) (CYPIIB6) (P450 IIB1). | |||||
|
CP2DB_MOUSE
|
||||||
| NC score | 0.750057 (rank : 81) | θ value | 1.80466e-16 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P24457 | Gene names | Cyp2d11, Cyp2d-11 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2D11 (EC 1.14.14.1) (CYPIID11) (P450-16-alpha) (CC) (Testosterone 16-alpha hydroxylase). | |||||
|
CP2F2_MOUSE
|
||||||
| NC score | 0.749542 (rank : 82) | θ value | 2.35696e-16 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P33267 | Gene names | Cyp2f2, Cyp2f-2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2F2 (EC 1.14.14.-) (CYPIIF2) (Naphthalene dehydrogenase) (Naphthalene hydroxylase) (P450-NAH-2). | |||||
|
CP2CJ_HUMAN
|
||||||
| NC score | 0.749226 (rank : 83) | θ value | 1.42992e-13 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P33261, P33259, Q8WZB1, Q8WZB2 | Gene names | CYP2C19 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C19 (EC 1.14.13.80) ((R)-limonene 6-monooxygenase) (EC 1.14.13.48) ((S)-limonene 6-monooxygenase) (EC 1.14.13.49) ((S)- limonene 7-monooxygenase) (CYPIIC19) (P450-11A) (Mephenytoin 4- hydroxylase) (CYPIIC17) (P450-254C). | |||||
|
CP2A6_HUMAN
|
||||||
| NC score | 0.749046 (rank : 84) | θ value | 1.80466e-16 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P11509, P00190, P10890, Q16803, Q4VAT9, Q4VAU0, Q4VAU1, Q9H1Z7, Q9UK48 | Gene names | CYP2A6 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2A6 (EC 1.14.14.1) (CYPIIA6) (Coumarin 7-hydroxylase) (P450 IIA3) (CYP2A3) (P450(I)). | |||||
|
CP2E1_MOUSE
|
||||||
| NC score | 0.748657 (rank : 85) | θ value | 1.86753e-13 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q05421, Q9Z198 | Gene names | Cyp2e1, Cyp2e, Cyp2e-1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2E1 (EC 1.14.14.1) (CYPIIE1) (P450-J) (P450-ALC). | |||||
|
CP2C8_HUMAN
|
||||||
| NC score | 0.744862 (rank : 86) | θ value | 4.16044e-13 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P10632 | Gene names | CYP2C8 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C8 (EC 1.14.14.1) (CYPIIC8) (P450 form 1) (P450 MP- 12/MP-20) (P450 IIC2) (S-mephenytoin 4-hydroxylase). | |||||
|
CP2A5_MOUSE
|
||||||
| NC score | 0.744827 (rank : 87) | θ value | 3.07829e-16 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P20852 | Gene names | Cyp2a5, Cyp2a-5 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2A5 (EC 1.14.14.1) (CYPIIA5) (Coumarin 7-hydroxylase) (P450-15-COH) (P450-IIA3.2). | |||||
|
CP2F1_HUMAN
|
||||||
| NC score | 0.744662 (rank : 88) | θ value | 1.68911e-14 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P24903 | Gene names | CYP2F1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2F1 (EC 1.14.14.1) (CYPIIF1). | |||||
|
CP2A7_HUMAN
|
||||||
| NC score | 0.743235 (rank : 89) | θ value | 2.35696e-16 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P20853, Q13121 | Gene names | CYP2A7 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2A7 (EC 1.14.14.1) (CYPIIA7) (P450-IIA4). | |||||
|
CP2CI_HUMAN
|
||||||
| NC score | 0.742894 (rank : 90) | θ value | 1.29331e-14 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P33260, Q16703, Q16751 | Gene names | CYP2C18 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C18 (EC 1.14.14.1) (CYPIIC18) (P450-6B/29C). | |||||
|
CP2A4_MOUSE
|
||||||
| NC score | 0.742732 (rank : 91) | θ value | 1.38178e-16 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P15392 | Gene names | Cyp2a4, Cyp2a-4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2A4 (EC 1.14.14.1) (CYPIIA4) (Testosterone 15-alpha- hydroxylase) (P450-15-alpha) (P450-IIA3.1). | |||||
|
CP2CT_MOUSE
|
||||||
| NC score | 0.739750 (rank : 92) | θ value | 1.16975e-15 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q64458, Q8WUN8 | Gene names | Cyp2c29 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C29 (EC 1.14.14.1) (CYPIIC29) (P-450 MUT-2) (Aldehyde oxygenase). | |||||
|
CP240_MOUSE
|
||||||
| NC score | 0.739698 (rank : 93) | θ value | 1.02475e-11 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P56657 | Gene names | Cyp2c40 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C40 (EC 1.14.14.1) (CYPIIC40). | |||||
|
CP2AC_MOUSE
|
||||||
| NC score | 0.739590 (rank : 94) | θ value | 1.16975e-15 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P56593 | Gene names | Cyp2a12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2A12 (EC 1.14.14.1) (CYPIIA12) (Steroid hormones 7- alpha-hydroxylase) (Testosterone 7-alpha-hydroxylase). | |||||
|
CP2BA_MOUSE
|
||||||
| NC score | 0.739167 (rank : 95) | θ value | 2.60593e-15 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | P12791 | Gene names | Cyp2b10, Cyp2b-10 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2B10 (EC 1.14.14.1) (CYPIIB10) (Testosterone 16-alpha hydroxylase) (P450-16-alpha) (Clone PF3/46). | |||||
|
CP237_MOUSE
|
||||||
| NC score | 0.738399 (rank : 96) | θ value | 1.2105e-12 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P56654 | Gene names | Cyp2c37 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C37 (EC 1.14.14.1) (CYPIIC37). | |||||
|
CP2AD_HUMAN
|
||||||
| NC score | 0.738037 (rank : 97) | θ value | 6.85773e-16 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q16696, Q53YR8, Q9H2X2 | Gene names | CYP2A13 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2A13 (EC 1.14.14.1) (CYPIIA13). | |||||
|
CP238_MOUSE
|
||||||
| NC score | 0.737238 (rank : 98) | θ value | 8.38298e-14 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P56655 | Gene names | Cyp2c38 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C38 (EC 1.14.14.1) (CYPIIC38). | |||||
|
CP7B1_HUMAN
|
||||||
| NC score | 0.647580 (rank : 99) | θ value | 1.99992e-07 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | O75881, Q9UNF5 | Gene names | CYP7B1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 7B1 (EC 1.14.13.100) (25-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase). | |||||
|
CP7B1_MOUSE
|
||||||
| NC score | 0.644676 (rank : 100) | θ value | 9.92553e-07 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q60991 | Gene names | Cyp7b1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 7B1 (EC 1.14.13.100) (25-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (HCT-1). | |||||
|
CP7A1_HUMAN
|
||||||
| NC score | 0.625202 (rank : 101) | θ value | 1.38499e-08 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P22680, P78454, Q7KZ19 | Gene names | CYP7A1, CYP7 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 7A1 (Cholesterol 7-alpha-monooxygenase) (CYPVII) (EC 1.14.13.17) (Cholesterol 7-alpha-hydroxylase). | |||||
|
CP7A1_MOUSE
|
||||||
| NC score | 0.621004 (rank : 102) | θ value | 6.87365e-08 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q64505 | Gene names | Cyp7a1, Cyp7 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 7A1 (EC 1.14.13.17) (Cholesterol 7-alpha- monooxygenase) (CYPVII) (Cholesterol 7-alpha-hydroxylase). | |||||
|
CP8B1_HUMAN
|
||||||
| NC score | 0.577925 (rank : 103) | θ value | 3.77169e-06 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9UNU6, O75958 | Gene names | CYP8B1, CYP12 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 8B1 (EC 1.14.13.95) (CYPVIIIB1) (7-alpha- hydroxycholest-4-en-3-one 12-alpha-hydroxylase) (Sterol 12-alpha- hydroxylase) (7-alpha-hydroxy-4-cholesten-3-one 12-alpha-hydroxylase). | |||||
|
PTGIS_MOUSE
|
||||||
| NC score | 0.574171 (rank : 104) | θ value | 5.44631e-05 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | O35074 | Gene names | Ptgis, Cyp8 | |||
|
Domain Architecture |

|
|||||
| Description | Prostacyclin synthase (EC 5.3.99.4) (Prostaglandin I2 synthase). | |||||
|
CP8B1_MOUSE
|
||||||
| NC score | 0.550576 (rank : 105) | θ value | 0.000461057 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O88962, Q9R217 | Gene names | Cyp8b1, Cyp12 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 8B1 (EC 1.14.13.95) (CYPVIIIB1) (7-alpha- hydroxycholest-4-en-3-one 12-alpha-hydroxylase) (Sterol 12-alpha- hydroxylase) (7-alpha-hydroxy-4-cholesten-3-one 12-alpha-hydroxylase). | |||||
|
PTGIS_HUMAN
|
||||||
| NC score | 0.508017 (rank : 106) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q16647, Q9HAX2, Q9HAX3, Q9HAX4 | Gene names | PTGIS, CYP8, CYP8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Prostacyclin synthase (EC 5.3.99.4) (Prostaglandin I2 synthase). | |||||
|
CENPK_HUMAN
|
||||||
| NC score | 0.015122 (rank : 107) | θ value | 0.125558 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BS16, Q9H4L0 | Gene names | CENPK, ICEN37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein K (CENP-K) (Interphase centromere complex protein 37) (Protein AF-5alpha) (p33). | |||||
|
UBE4B_MOUSE
|
||||||
| NC score | 0.010246 (rank : 108) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ES00, Q9EQE0 | Gene names | Ube4b, Ufd2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Ufd2a). | |||||
|
UBE4B_HUMAN
|
||||||
| NC score | 0.009146 (rank : 109) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
COX3_HUMAN
|
||||||
| NC score | 0.006578 (rank : 110) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00414 | Gene names | MT-CO3, COIII, COXIII, MTCO3 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome c oxidase subunit 3 (EC 1.9.3.1) (Cytochrome c oxidase polypeptide III). | |||||
|
DHC24_HUMAN
|
||||||
| NC score | 0.006416 (rank : 111) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15392, Q9HBA8 | Gene names | DHCR24, KIAA0018 | |||
|
Domain Architecture |

|
|||||
| Description | 24-dehydrocholesterol reductase precursor (EC 1.3.1.-) (3-beta- hydroxysterol delta-24-reductase) (Seladin-1) (Diminuto/dwarf1 homolog). | |||||
|
SPAT7_HUMAN
|
||||||
| NC score | 0.005370 (rank : 112) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P0W8, Q8WX30, Q96HF3, Q9H0X0, Q9P0W7 | Gene names | SPATA7, HSD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 7 (Spermatogenesis-associated protein HSD3) (HSD-3.1). | |||||
|
AXUD1_MOUSE
|
||||||
| NC score | 0.004637 (rank : 113) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59054 | Gene names | Axud1, Taip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axin-1 up-regulated gene 1 protein (TGF-beta-induced apoptosis protein 3) (TAIP-3). | |||||
|
UB2Q2_MOUSE
|
||||||
| NC score | 0.002917 (rank : 114) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2Z8, Q3UBX3, Q3V3A5, Q8BUN2, Q8BVX5 | Gene names | Ube2q2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 Q2 (EC 6.3.2.19) (Ubiquitin-protein ligase Q2) (Ubiquitin carrier protein Q2). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | -0.001105 (rank : 115) | θ value | 1.81305 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | -0.001491 (rank : 116) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | -0.001890 (rank : 117) | θ value | 0.62314 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
STK10_MOUSE
|
||||||
| NC score | -0.011793 (rank : 118) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||