Please be patient as the page loads
|
CL005_HUMAN
|
||||||
| SwissProt Accessions | Q9NQ88 | Gene names | C12orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf5. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CL005_HUMAN
|
||||||
| θ value | 4.56552e-153 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NQ88 | Gene names | C12orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf5. | |||||
|
CL005_MOUSE
|
||||||
| θ value | 1.25781e-110 (rank : 2) | NC score | 0.992801 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BZA9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf5 homolog. | |||||
|
PMGE_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 3) | NC score | 0.278138 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15327, Q543Z6 | Gene names | Bpgm | |||
|
Domain Architecture |
|
|||||
| Description | Bisphosphoglycerate mutase (EC 5.4.2.4) (2,3-bisphosphoglycerate mutase, erythrocyte) (2,3-bisphosphoglycerate synthase) (BPGM) (EC 5.4.2.1) (EC 3.1.3.13) (BPG-dependent PGAM). | |||||
|
PGAM2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 4) | NC score | 0.260111 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15259 | Gene names | PGAM2, PGAMM | |||
|
Domain Architecture |
|
|||||
| Description | Phosphoglycerate mutase 2 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme M) (PGAM-M) (BPG-dependent PGAM 2) (Muscle-specific phosphoglycerate mutase). | |||||
|
PGAM2_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 5) | NC score | 0.260410 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70250 | Gene names | Pgam2 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphoglycerate mutase 2 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme M) (PGAM-M) (BPG-dependent PGAM 2) (Muscle-specific phosphoglycerate mutase). | |||||
|
PMGE_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 6) | NC score | 0.267951 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P07738 | Gene names | BPGM | |||
|
Domain Architecture |
|
|||||
| Description | Bisphosphoglycerate mutase (EC 5.4.2.4) (2,3-bisphosphoglycerate mutase, erythrocyte) (2,3-bisphosphoglycerate synthase) (BPGM) (EC 5.4.2.1) (EC 3.1.3.13) (BPG-dependent PGAM). | |||||
|
PGAM1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.248830 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P18669, Q9BWC0 | Gene names | PGAM1 | |||
|
Domain Architecture |
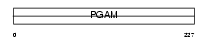
|
|||||
| Description | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1). | |||||
|
PGAM1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.248878 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9DBJ1, Q9D6W0, Q9ERT3 | Gene names | Pgam1 | |||
|
Domain Architecture |
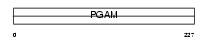
|
|||||
| Description | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1). | |||||
|
PGAM4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 9) | NC score | 0.225755 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N0Y7, Q8NI24, Q8NI25, Q8NI26 | Gene names | PGAM4, PGAM3 | |||
|
Domain Architecture |
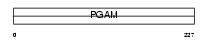
|
|||||
| Description | Probable phosphoglycerate mutase 4 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13). | |||||
|
STS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 10) | NC score | 0.035392 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57075, Q6HA34, Q6HA35, Q6ISI6, Q6ISK3, Q6ISS9 | Gene names | UBASH3A, STS2 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of T-cell receptor signaling 2 (STS-2) (Cbl-interacting protein 4) (CLIP4) (T-cell ubiquitin ligand) (TULA). | |||||
|
RPC5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 11) | NC score | 0.024976 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZT4, Q8CI35, Q9DBD1 | Gene names | Polr3e, Sin | |||
|
Domain Architecture |
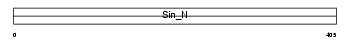
|
|||||
| Description | DNA-directed RNA polymerases III 80 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 5) (RPC5) (Sex-lethal interactor homolog). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.004147 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CL005_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.56552e-153 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NQ88 | Gene names | C12orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf5. | |||||
|
CL005_MOUSE
|
||||||
| NC score | 0.992801 (rank : 2) | θ value | 1.25781e-110 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BZA9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf5 homolog. | |||||
|
PMGE_MOUSE
|
||||||
| NC score | 0.278138 (rank : 3) | θ value | 0.00175202 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15327, Q543Z6 | Gene names | Bpgm | |||
|
Domain Architecture |

|
|||||
| Description | Bisphosphoglycerate mutase (EC 5.4.2.4) (2,3-bisphosphoglycerate mutase, erythrocyte) (2,3-bisphosphoglycerate synthase) (BPGM) (EC 5.4.2.1) (EC 3.1.3.13) (BPG-dependent PGAM). | |||||
|
PMGE_HUMAN
|
||||||
| NC score | 0.267951 (rank : 4) | θ value | 0.0113563 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P07738 | Gene names | BPGM | |||
|
Domain Architecture |

|
|||||
| Description | Bisphosphoglycerate mutase (EC 5.4.2.4) (2,3-bisphosphoglycerate mutase, erythrocyte) (2,3-bisphosphoglycerate synthase) (BPGM) (EC 5.4.2.1) (EC 3.1.3.13) (BPG-dependent PGAM). | |||||
|
PGAM2_MOUSE
|
||||||
| NC score | 0.260410 (rank : 5) | θ value | 0.00665767 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70250 | Gene names | Pgam2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoglycerate mutase 2 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme M) (PGAM-M) (BPG-dependent PGAM 2) (Muscle-specific phosphoglycerate mutase). | |||||
|
PGAM2_HUMAN
|
||||||
| NC score | 0.260111 (rank : 6) | θ value | 0.00665767 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15259 | Gene names | PGAM2, PGAMM | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoglycerate mutase 2 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme M) (PGAM-M) (BPG-dependent PGAM 2) (Muscle-specific phosphoglycerate mutase). | |||||
|
PGAM1_MOUSE
|
||||||
| NC score | 0.248878 (rank : 7) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9DBJ1, Q9D6W0, Q9ERT3 | Gene names | Pgam1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1). | |||||
|
PGAM1_HUMAN
|
||||||
| NC score | 0.248830 (rank : 8) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P18669, Q9BWC0 | Gene names | PGAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1). | |||||
|
PGAM4_HUMAN
|
||||||
| NC score | 0.225755 (rank : 9) | θ value | 1.06291 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N0Y7, Q8NI24, Q8NI25, Q8NI26 | Gene names | PGAM4, PGAM3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phosphoglycerate mutase 4 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13). | |||||
|
STS2_HUMAN
|
||||||
| NC score | 0.035392 (rank : 10) | θ value | 4.03905 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57075, Q6HA34, Q6HA35, Q6ISI6, Q6ISK3, Q6ISS9 | Gene names | UBASH3A, STS2 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of T-cell receptor signaling 2 (STS-2) (Cbl-interacting protein 4) (CLIP4) (T-cell ubiquitin ligand) (TULA). | |||||
|
RPC5_MOUSE
|
||||||
| NC score | 0.024976 (rank : 11) | θ value | 5.27518 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZT4, Q8CI35, Q9DBD1 | Gene names | Polr3e, Sin | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerases III 80 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 5) (RPC5) (Sex-lethal interactor homolog). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.004147 (rank : 12) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||