Please be patient as the page loads
|
CHST9_HUMAN
|
||||||
| SwissProt Accessions | Q7L1S5, Q6UX69, Q9BXH3, Q9BXH4, Q9BZW9 | Gene names | CHST9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CHST9_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7L1S5, Q6UX69, Q9BXH3, Q9BXH4, Q9BZW9 | Gene names | CHST9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
|
CHST9_MOUSE
|
||||||
| θ value | 3.24166e-183 (rank : 2) | NC score | 0.997512 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q76EC5 | Gene names | Chst9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
|
CHST8_MOUSE
|
||||||
| θ value | 6.24247e-110 (rank : 3) | NC score | 0.984728 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BQ86, Q76EC6, Q80XD4 | Gene names | Chst8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
|
CHST8_HUMAN
|
||||||
| θ value | 1.53757e-108 (rank : 4) | NC score | 0.985171 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H2A9, Q9H3N2 | Gene names | CHST8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
|
CHSTB_HUMAN
|
||||||
| θ value | 7.49467e-55 (rank : 5) | NC score | 0.939011 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NPF2, Q9NXY6, Q9NY36 | Gene names | CHST11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
CHSTB_MOUSE
|
||||||
| θ value | 7.49467e-55 (rank : 6) | NC score | 0.938920 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JME2, Q9JJS2 | Gene names | Chst11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
CHSTD_HUMAN
|
||||||
| θ value | 4.2656e-42 (rank : 7) | NC score | 0.939519 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NET6 | Gene names | CHST13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 13 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 3) (Chondroitin 4-sulfotransferase 3) (C4ST3) (C4ST- 3). | |||||
|
CHSTC_HUMAN
|
||||||
| θ value | 1.79215e-40 (rank : 8) | NC score | 0.913335 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRB3, Q9NXY7 | Gene names | CHST12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2) (Sulfotransferase Hlo). | |||||
|
CHSTC_MOUSE
|
||||||
| θ value | 3.05695e-40 (rank : 9) | NC score | 0.909898 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99LL3, Q9JJS3 | Gene names | Chst12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2). | |||||
|
CHSTA_MOUSE
|
||||||
| θ value | 3.99248e-40 (rank : 10) | NC score | 0.905898 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PGK7, Q8BKU3, Q8BL08, Q8R2Y5, Q91Y40 | Gene names | Chst10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST). | |||||
|
CHSTA_HUMAN
|
||||||
| θ value | 3.73686e-38 (rank : 11) | NC score | 0.903657 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43529 | Gene names | CHST10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST) (huHNK-1ST). | |||||
|
CHSTE_MOUSE
|
||||||
| θ value | 2.26708e-35 (rank : 12) | NC score | 0.899296 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80V53, Q3TXA1, Q8R304, Q9D0P2 | Gene names | D4st1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase D4ST1 (EC 2.8.2.-) (Dermatan 4- sulfotransferase 1) (D4ST-1). | |||||
|
CHSTE_HUMAN
|
||||||
| θ value | 1.9192e-34 (rank : 13) | NC score | 0.898850 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NCH0, Q6PJ31, Q6UXA0, Q96P94 | Gene names | D4ST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase D4ST1 (EC 2.8.2.-) (Dermatan 4- sulfotransferase 1) (D4ST-1) (hD4ST). | |||||
|
RLF_HUMAN
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.010010 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13129, Q9NU60 | Gene names | RLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Rlf (Rearranged L-myc fusion gene protein) (Zn-15- related protein). | |||||
|
ERCC3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.019377 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19447 | Gene names | ERCC3, XPB, XPBC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase XPB subunit (EC 3.6.1.-) (Basic transcription factor 2 89 kDa subunit) (BTF2-p89) (TFIIH 89 kDa subunit) (DNA-repair protein complementing XP-B cells) (Xeroderma pigmentosum group B-complementing protein) (DNA excision repair protein ERCC-3). | |||||
|
TEKT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.011181 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969V4 | Gene names | TEKT1 | |||
|
Domain Architecture |

|
|||||
| Description | Tektin-1. | |||||
|
K2027_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.016377 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
TES_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.014549 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47226 | Gene names | Tes | |||
|
Domain Architecture |
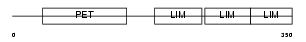
|
|||||
| Description | Testin (TES1/TES2). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.002369 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.007189 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.000469 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
ITA9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.003350 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13797, Q14638 | Gene names | ITGA9 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-9 precursor (Integrin alpha-RLC). | |||||
|
NOL8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.006677 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
CHST9_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7L1S5, Q6UX69, Q9BXH3, Q9BXH4, Q9BZW9 | Gene names | CHST9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
|
CHST9_MOUSE
|
||||||
| NC score | 0.997512 (rank : 2) | θ value | 3.24166e-183 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q76EC5 | Gene names | Chst9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
|
CHST8_HUMAN
|
||||||
| NC score | 0.985171 (rank : 3) | θ value | 1.53757e-108 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H2A9, Q9H3N2 | Gene names | CHST8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
|
CHST8_MOUSE
|
||||||
| NC score | 0.984728 (rank : 4) | θ value | 6.24247e-110 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BQ86, Q76EC6, Q80XD4 | Gene names | Chst8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
|
CHSTD_HUMAN
|
||||||
| NC score | 0.939519 (rank : 5) | θ value | 4.2656e-42 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NET6 | Gene names | CHST13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 13 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 3) (Chondroitin 4-sulfotransferase 3) (C4ST3) (C4ST- 3). | |||||
|
CHSTB_HUMAN
|
||||||
| NC score | 0.939011 (rank : 6) | θ value | 7.49467e-55 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NPF2, Q9NXY6, Q9NY36 | Gene names | CHST11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
CHSTB_MOUSE
|
||||||
| NC score | 0.938920 (rank : 7) | θ value | 7.49467e-55 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JME2, Q9JJS2 | Gene names | Chst11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
CHSTC_HUMAN
|
||||||
| NC score | 0.913335 (rank : 8) | θ value | 1.79215e-40 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRB3, Q9NXY7 | Gene names | CHST12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2) (Sulfotransferase Hlo). | |||||
|
CHSTC_MOUSE
|
||||||
| NC score | 0.909898 (rank : 9) | θ value | 3.05695e-40 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99LL3, Q9JJS3 | Gene names | Chst12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2). | |||||
|
CHSTA_MOUSE
|
||||||
| NC score | 0.905898 (rank : 10) | θ value | 3.99248e-40 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PGK7, Q8BKU3, Q8BL08, Q8R2Y5, Q91Y40 | Gene names | Chst10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST). | |||||
|
CHSTA_HUMAN
|
||||||
| NC score | 0.903657 (rank : 11) | θ value | 3.73686e-38 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43529 | Gene names | CHST10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST) (huHNK-1ST). | |||||
|
CHSTE_MOUSE
|
||||||
| NC score | 0.899296 (rank : 12) | θ value | 2.26708e-35 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80V53, Q3TXA1, Q8R304, Q9D0P2 | Gene names | D4st1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase D4ST1 (EC 2.8.2.-) (Dermatan 4- sulfotransferase 1) (D4ST-1). | |||||
|
CHSTE_HUMAN
|
||||||
| NC score | 0.898850 (rank : 13) | θ value | 1.9192e-34 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NCH0, Q6PJ31, Q6UXA0, Q96P94 | Gene names | D4ST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase D4ST1 (EC 2.8.2.-) (Dermatan 4- sulfotransferase 1) (D4ST-1) (hD4ST). | |||||
|
ERCC3_HUMAN
|
||||||
| NC score | 0.019377 (rank : 14) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19447 | Gene names | ERCC3, XPB, XPBC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase XPB subunit (EC 3.6.1.-) (Basic transcription factor 2 89 kDa subunit) (BTF2-p89) (TFIIH 89 kDa subunit) (DNA-repair protein complementing XP-B cells) (Xeroderma pigmentosum group B-complementing protein) (DNA excision repair protein ERCC-3). | |||||
|
K2027_HUMAN
|
||||||
| NC score | 0.016377 (rank : 15) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
TES_MOUSE
|
||||||
| NC score | 0.014549 (rank : 16) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47226 | Gene names | Tes | |||
|
Domain Architecture |

|
|||||
| Description | Testin (TES1/TES2). | |||||
|
TEKT1_HUMAN
|
||||||
| NC score | 0.011181 (rank : 17) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969V4 | Gene names | TEKT1 | |||
|
Domain Architecture |

|
|||||
| Description | Tektin-1. | |||||
|
RLF_HUMAN
|
||||||
| NC score | 0.010010 (rank : 18) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13129, Q9NU60 | Gene names | RLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Rlf (Rearranged L-myc fusion gene protein) (Zn-15- related protein). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.007189 (rank : 19) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
NOL8_HUMAN
|
||||||
| NC score | 0.006677 (rank : 20) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
ITA9_HUMAN
|
||||||
| NC score | 0.003350 (rank : 21) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13797, Q14638 | Gene names | ITGA9 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-9 precursor (Integrin alpha-RLC). | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.002369 (rank : 22) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.000469 (rank : 23) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||