Please be patient as the page loads
|
CATB_HUMAN
|
||||||
| SwissProt Accessions | P07858, Q96D87 | Gene names | CTSB, CPSB | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) (APP secretase) (APPS) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CATB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P07858, Q96D87 | Gene names | CTSB, CPSB | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) (APP secretase) (APPS) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
|
CATB_MOUSE
|
||||||
| θ value | 1.36511e-173 (rank : 2) | NC score | 0.998022 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10605, Q3UDW7 | Gene names | Ctsb | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
|
TINAL_HUMAN
|
||||||
| θ value | 9.83387e-39 (rank : 3) | NC score | 0.887960 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9GZM7, Q8TEJ9, Q8WZ23, Q96GZ4, Q96JW3 | Gene names | TINAGL1, GIS5, LCN7, OLRG2, TINAGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Tubulointerstitial nephritis antigen-related protein) (TIN Ag-related protein) (TIN-Ag-RP) (Glucocorticoid-inducible protein 5) (Oxidized LDL-responsive gene 2 protein) (OLRG-2). | |||||
|
TINAL_MOUSE
|
||||||
| θ value | 4.13158e-37 (rank : 4) | NC score | 0.884025 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
TINAG_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 5) | NC score | 0.875935 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |
|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
CATC_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 6) | NC score | 0.897788 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P53634, Q8WYA7, Q8WYA8 | Gene names | CTSC, CPPI | |||
|
Domain Architecture |
|
|||||
| Description | Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl- peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)]. | |||||
|
CATC_MOUSE
|
||||||
| θ value | 7.80994e-28 (rank : 7) | NC score | 0.894953 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97821, O08853 | Gene names | Ctsc | |||
|
Domain Architecture |
|
|||||
| Description | Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl- peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)]. | |||||
|
CATH_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 8) | NC score | 0.868210 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49935 | Gene names | Ctsh | |||
|
Domain Architecture |
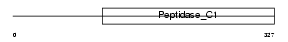
|
|||||
| Description | Cathepsin H precursor (EC 3.4.22.16) (Cathepsin B3) (Cathepsin BA) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain]. | |||||
|
CATH_HUMAN
|
||||||
| θ value | 1.37858e-24 (rank : 9) | NC score | 0.865995 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09668, Q9BUM7 | Gene names | CTSH | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin H precursor (EC 3.4.22.16) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain]. | |||||
|
CATS_HUMAN
|
||||||
| θ value | 2.59989e-23 (rank : 10) | NC score | 0.845463 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P25774, Q5T5I0, Q9BUG3 | Gene names | CTSS | |||
|
Domain Architecture |
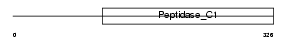
|
|||||
| Description | Cathepsin S precursor (EC 3.4.22.27). | |||||
|
CATS_MOUSE
|
||||||
| θ value | 5.79196e-23 (rank : 11) | NC score | 0.841843 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70370, O54973 | Gene names | Ctss, Cats | |||
|
Domain Architecture |
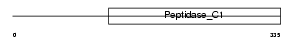
|
|||||
| Description | Cathepsin S precursor (EC 3.4.22.27). | |||||
|
CATL_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 12) | NC score | 0.824172 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07711, Q96QJ0 | Gene names | CTSL | |||
|
Domain Architecture |
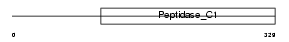
|
|||||
| Description | Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) [Contains: Cathepsin L heavy chain; Cathepsin L light chain]. | |||||
|
CATL2_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 13) | NC score | 0.825494 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60911, O60233, Q5T1U0 | Gene names | CTSL2, CATL2, CTSU, CTSV | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin L2 precursor (EC 3.4.22.43) (Cathepsin V) (Cathepsin U). | |||||
|
CATK_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 14) | NC score | 0.830807 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43235 | Gene names | CTSK, CTSO, CTSO2 | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin K precursor (EC 3.4.22.38) (Cathepsin O) (Cathepsin X) (Cathepsin O2). | |||||
|
CATZ_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 15) | NC score | 0.855396 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBR2, O75331, Q9UQV5, Q9UQV6 | Gene names | CTSZ | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-) (Cathepsin X) (Cathepsin P). | |||||
|
CATL_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 16) | NC score | 0.819345 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P06797, Q91UZ0 | Gene names | Ctsl | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) (p39 cysteine proteinase) [Contains: Cathepsin L heavy chain; Cathepsin L light chain]. | |||||
|
CATZ_MOUSE
|
||||||
| θ value | 4.29542e-18 (rank : 17) | NC score | 0.854976 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WUU7 | Gene names | Ctsz | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-). | |||||
|
CATF_HUMAN
|
||||||
| θ value | 9.56915e-18 (rank : 18) | NC score | 0.816243 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBX1, O95240, Q9NSU4, Q9UKQ5 | Gene names | CTSF | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin F precursor (EC 3.4.22.41) (CATSF). | |||||
|
CATO_HUMAN
|
||||||
| θ value | 9.56915e-18 (rank : 19) | NC score | 0.845074 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43234 | Gene names | CTSO, CTSO1 | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin O precursor (EC 3.4.22.42). | |||||
|
CATF_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 20) | NC score | 0.816507 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R013, Q9WUT4 | Gene names | Ctsf | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin F precursor (EC 3.4.22.41). | |||||
|
CATJ_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 21) | NC score | 0.822982 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R014, Q564D9, Q91XK6, Q9WV51 | Gene names | Ctsj, Ctsp | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin J precursor (EC 3.4.22.-) (Cathepsin P) (Cathepsin L-related protein) (Catlrp-p). | |||||
|
CATK_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 22) | NC score | 0.828939 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55097, O88718 | Gene names | Ctsk | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin K precursor (EC 3.4.22.38). | |||||
|
CATO_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 23) | NC score | 0.837708 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BM88, Q3TM69, Q80V35, Q8BKX0 | Gene names | Ctso | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cathepsin O precursor (EC 3.4.22.42). | |||||
|
CATR_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 24) | NC score | 0.814204 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JIA9 | Gene names | Ctsr, Catr | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin R precursor (EC 3.4.22.-). | |||||
|
CATW_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 25) | NC score | 0.816005 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56203 | Gene names | Ctsw | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin W precursor (EC 3.4.22.-) (Lymphopain). | |||||
|
CATM_MOUSE
|
||||||
| θ value | 9.26847e-13 (rank : 26) | NC score | 0.793359 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JL96, Q91Z75, Q91ZF3, Q9CQB9 | Gene names | Ctsm, Catm | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin M precursor (EC 3.4.22.-). | |||||
|
CATW_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 27) | NC score | 0.810603 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56202, Q86VT4 | Gene names | CTSW | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin W precursor (EC 3.4.22.-) (Lymphopain). | |||||
|
DUX4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.015703 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBX2 | Gene names | DUX4, DUX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double homeobox protein 4 (Double homeobox protein 10) (Double homeobox protein 4/10). | |||||
|
PDZD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.007685 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
ZN282_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | -0.002468 (rank : 39) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UDV7, O43691, Q6DKK0 | Gene names | ZNF282, HUB1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 282 (HTLV-I U5RE-binding protein 1) (HUB-1). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.004477 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
THOC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.023688 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96FV9, Q15219 | Gene names | THOC1, HPR1 | |||
|
Domain Architecture |

|
|||||
| Description | THO complex subunit 1 (Tho1) (Nuclear matrix protein p84). | |||||
|
THOC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.023651 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3N6, Q8BWD5, Q8BXY3 | Gene names | Thoc1 | |||
|
Domain Architecture |

|
|||||
| Description | THO complex subunit 1 (Tho1) (Nuclear matrix protein p84). | |||||
|
CDK10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | -0.001829 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15131, Q15130 | Gene names | CDK10 | |||
|
Domain Architecture |
|
|||||
| Description | Cell division protein kinase 10 (EC 2.7.11.22) (Serine/threonine- protein kinase PISSLRE). | |||||
|
LAMA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.003182 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
EGFR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | -0.002745 (rank : 40) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 927 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01279 | Gene names | Egfr | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1). | |||||
|
RELN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.006511 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |
|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
STAB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.001143 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
CTL2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.440071 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12399 | Gene names | Ctla2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTLA-2-alpha protein precursor. | |||||
|
TSP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.268700 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13438 | Gene names | Tpbpa, Tpbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophoblast-specific protein alpha precursor. | |||||
|
CATB_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P07858, Q96D87 | Gene names | CTSB, CPSB | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) (APP secretase) (APPS) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
|
CATB_MOUSE
|
||||||
| NC score | 0.998022 (rank : 2) | θ value | 1.36511e-173 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10605, Q3UDW7 | Gene names | Ctsb | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
|
CATC_HUMAN
|
||||||
| NC score | 0.897788 (rank : 3) | θ value | 2.19584e-30 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P53634, Q8WYA7, Q8WYA8 | Gene names | CTSC, CPPI | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl- peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)]. | |||||
|
CATC_MOUSE
|
||||||
| NC score | 0.894953 (rank : 4) | θ value | 7.80994e-28 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97821, O08853 | Gene names | Ctsc | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl- peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)]. | |||||
|
TINAL_HUMAN
|
||||||
| NC score | 0.887960 (rank : 5) | θ value | 9.83387e-39 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9GZM7, Q8TEJ9, Q8WZ23, Q96GZ4, Q96JW3 | Gene names | TINAGL1, GIS5, LCN7, OLRG2, TINAGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Tubulointerstitial nephritis antigen-related protein) (TIN Ag-related protein) (TIN-Ag-RP) (Glucocorticoid-inducible protein 5) (Oxidized LDL-responsive gene 2 protein) (OLRG-2). | |||||
|
TINAL_MOUSE
|
||||||
| NC score | 0.884025 (rank : 6) | θ value | 4.13158e-37 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
TINAG_HUMAN
|
||||||
| NC score | 0.875935 (rank : 7) | θ value | 3.06405e-32 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |

|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
CATH_MOUSE
|
||||||
| NC score | 0.868210 (rank : 8) | θ value | 1.05554e-24 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49935 | Gene names | Ctsh | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin H precursor (EC 3.4.22.16) (Cathepsin B3) (Cathepsin BA) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain]. | |||||
|
CATH_HUMAN
|
||||||
| NC score | 0.865995 (rank : 9) | θ value | 1.37858e-24 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09668, Q9BUM7 | Gene names | CTSH | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin H precursor (EC 3.4.22.16) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain]. | |||||
|
CATZ_HUMAN
|
||||||
| NC score | 0.855396 (rank : 10) | θ value | 1.47631e-18 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBR2, O75331, Q9UQV5, Q9UQV6 | Gene names | CTSZ | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-) (Cathepsin X) (Cathepsin P). | |||||
|
CATZ_MOUSE
|
||||||
| NC score | 0.854976 (rank : 11) | θ value | 4.29542e-18 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WUU7 | Gene names | Ctsz | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-). | |||||
|
CATS_HUMAN
|
||||||
| NC score | 0.845463 (rank : 12) | θ value | 2.59989e-23 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P25774, Q5T5I0, Q9BUG3 | Gene names | CTSS | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin S precursor (EC 3.4.22.27). | |||||
|
CATO_HUMAN
|
||||||
| NC score | 0.845074 (rank : 13) | θ value | 9.56915e-18 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43234 | Gene names | CTSO, CTSO1 | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin O precursor (EC 3.4.22.42). | |||||
|
CATS_MOUSE
|
||||||
| NC score | 0.841843 (rank : 14) | θ value | 5.79196e-23 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70370, O54973 | Gene names | Ctss, Cats | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin S precursor (EC 3.4.22.27). | |||||
|
CATO_MOUSE
|
||||||
| NC score | 0.837708 (rank : 15) | θ value | 1.16975e-15 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BM88, Q3TM69, Q80V35, Q8BKX0 | Gene names | Ctso | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cathepsin O precursor (EC 3.4.22.42). | |||||
|
CATK_HUMAN
|
||||||
| NC score | 0.830807 (rank : 16) | θ value | 2.97466e-19 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43235 | Gene names | CTSK, CTSO, CTSO2 | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin K precursor (EC 3.4.22.38) (Cathepsin O) (Cathepsin X) (Cathepsin O2). | |||||
|
CATK_MOUSE
|
||||||
| NC score | 0.828939 (rank : 17) | θ value | 3.63628e-17 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55097, O88718 | Gene names | Ctsk | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin K precursor (EC 3.4.22.38). | |||||
|
CATL2_HUMAN
|
||||||
| NC score | 0.825494 (rank : 18) | θ value | 2.69047e-20 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60911, O60233, Q5T1U0 | Gene names | CTSL2, CATL2, CTSU, CTSV | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin L2 precursor (EC 3.4.22.43) (Cathepsin V) (Cathepsin U). | |||||
|
CATL_HUMAN
|
||||||
| NC score | 0.824172 (rank : 19) | θ value | 4.15078e-21 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07711, Q96QJ0 | Gene names | CTSL | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) [Contains: Cathepsin L heavy chain; Cathepsin L light chain]. | |||||
|
CATJ_MOUSE
|
||||||
| NC score | 0.822982 (rank : 20) | θ value | 2.13179e-17 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R014, Q564D9, Q91XK6, Q9WV51 | Gene names | Ctsj, Ctsp | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin J precursor (EC 3.4.22.-) (Cathepsin P) (Cathepsin L-related protein) (Catlrp-p). | |||||
|
CATL_MOUSE
|
||||||
| NC score | 0.819345 (rank : 21) | θ value | 1.92812e-18 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P06797, Q91UZ0 | Gene names | Ctsl | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) (p39 cysteine proteinase) [Contains: Cathepsin L heavy chain; Cathepsin L light chain]. | |||||
|
CATF_MOUSE
|
||||||
| NC score | 0.816507 (rank : 22) | θ value | 2.13179e-17 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R013, Q9WUT4 | Gene names | Ctsf | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin F precursor (EC 3.4.22.41). | |||||
|
CATF_HUMAN
|
||||||
| NC score | 0.816243 (rank : 23) | θ value | 9.56915e-18 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBX1, O95240, Q9NSU4, Q9UKQ5 | Gene names | CTSF | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin F precursor (EC 3.4.22.41) (CATSF). | |||||
|
CATW_MOUSE
|
||||||
| NC score | 0.816005 (rank : 24) | θ value | 3.18553e-13 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56203 | Gene names | Ctsw | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin W precursor (EC 3.4.22.-) (Lymphopain). | |||||
|
CATR_MOUSE
|
||||||
| NC score | 0.814204 (rank : 25) | θ value | 7.58209e-15 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JIA9 | Gene names | Ctsr, Catr | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin R precursor (EC 3.4.22.-). | |||||
|
CATW_HUMAN
|
||||||
| NC score | 0.810603 (rank : 26) | θ value | 9.26847e-13 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56202, Q86VT4 | Gene names | CTSW | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin W precursor (EC 3.4.22.-) (Lymphopain). | |||||
|
CATM_MOUSE
|
||||||
| NC score | 0.793359 (rank : 27) | θ value | 9.26847e-13 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JL96, Q91Z75, Q91ZF3, Q9CQB9 | Gene names | Ctsm, Catm | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin M precursor (EC 3.4.22.-). | |||||
|
CTL2A_MOUSE
|
||||||
| NC score | 0.440071 (rank : 28) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12399 | Gene names | Ctla2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTLA-2-alpha protein precursor. | |||||
|
TSP_MOUSE
|
||||||
| NC score | 0.268700 (rank : 29) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13438 | Gene names | Tpbpa, Tpbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophoblast-specific protein alpha precursor. | |||||
|
THOC1_HUMAN
|
||||||
| NC score | 0.023688 (rank : 30) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96FV9, Q15219 | Gene names | THOC1, HPR1 | |||
|
Domain Architecture |

|
|||||
| Description | THO complex subunit 1 (Tho1) (Nuclear matrix protein p84). | |||||
|
THOC1_MOUSE
|
||||||
| NC score | 0.023651 (rank : 31) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3N6, Q8BWD5, Q8BXY3 | Gene names | Thoc1 | |||
|
Domain Architecture |

|
|||||
| Description | THO complex subunit 1 (Tho1) (Nuclear matrix protein p84). | |||||
|
DUX4_HUMAN
|
||||||
| NC score | 0.015703 (rank : 32) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBX2 | Gene names | DUX4, DUX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double homeobox protein 4 (Double homeobox protein 10) (Double homeobox protein 4/10). | |||||
|
PDZD1_MOUSE
|
||||||
| NC score | 0.007685 (rank : 33) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
RELN_HUMAN
|
||||||
| NC score | 0.006511 (rank : 34) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |

|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.004477 (rank : 35) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
LAMA1_MOUSE
|
||||||
| NC score | 0.003182 (rank : 36) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.001143 (rank : 37) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
CDK10_HUMAN
|
||||||
| NC score | -0.001829 (rank : 38) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15131, Q15130 | Gene names | CDK10 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division protein kinase 10 (EC 2.7.11.22) (Serine/threonine- protein kinase PISSLRE). | |||||
|
ZN282_HUMAN
|
||||||
| NC score | -0.002468 (rank : 39) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UDV7, O43691, Q6DKK0 | Gene names | ZNF282, HUB1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 282 (HTLV-I U5RE-binding protein 1) (HUB-1). | |||||
|
EGFR_MOUSE
|
||||||
| NC score | -0.002745 (rank : 40) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 927 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01279 | Gene names | Egfr | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1). | |||||