Please be patient as the page loads
|
BACE2_HUMAN
|
||||||
| SwissProt Accessions | Q9Y5Z0, Q9UJT6 | Gene names | BACE2, ASP21 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-secretase 2 precursor (EC 3.4.23.45) (Beta-site APP-cleaving enzyme 2) (Aspartyl protease 1) (Asp 1) (ASP1) (Membrane-associated aspartic protease 1) (Memapsin-1) (Down region aspartic protease). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
BACE2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5Z0, Q9UJT6 | Gene names | BACE2, ASP21 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-secretase 2 precursor (EC 3.4.23.45) (Beta-site APP-cleaving enzyme 2) (Aspartyl protease 1) (Asp 1) (ASP1) (Membrane-associated aspartic protease 1) (Memapsin-1) (Down region aspartic protease). | |||||
|
BACE1_HUMAN
|
||||||
| θ value | 3.29472e-127 (rank : 2) | NC score | 0.976835 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56817, Q9BYB9, Q9BYC0, Q9BYC1, Q9UJT5 | Gene names | BACE1, BACE | |||
|
Domain Architecture |
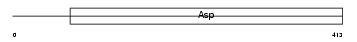
|
|||||
| Description | Beta-secretase 1 precursor (EC 3.4.23.46) (Beta-site APP cleaving enzyme 1) (Beta-site amyloid precursor protein cleaving enzyme 1) (Membrane-associated aspartic protease 2) (Memapsin-2) (Aspartyl protease 2) (Asp 2) (ASP2). | |||||
|
BACE1_MOUSE
|
||||||
| θ value | 3.29472e-127 (rank : 3) | NC score | 0.977177 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56818, Q544D0 | Gene names | Bace1, Bace | |||
|
Domain Architecture |
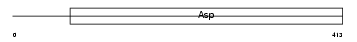
|
|||||
| Description | Beta-secretase 1 precursor (EC 3.4.23.46) (Beta-site APP cleaving enzyme 1) (Beta-site amyloid precursor protein cleaving enzyme 1) (Membrane-associated aspartic protease 2) (Memapsin-2) (Aspartyl protease 2) (Asp 2) (ASP2). | |||||
|
PEPC_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 4) | NC score | 0.810715 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P20142, Q5T3D7, Q5T3D8 | Gene names | PGC | |||
|
Domain Architecture |
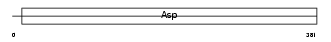
|
|||||
| Description | Gastricsin precursor (EC 3.4.23.3) (Pepsinogen C). | |||||
|
PEPC_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 5) | NC score | 0.801231 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D7R7, Q9D7T2 | Gene names | Pgc, Upg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gastricsin precursor (EC 3.4.23.3) (Pepsinogen C). | |||||
|
CATE_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 6) | NC score | 0.801693 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70269, O35647 | Gene names | Ctse | |||
|
Domain Architecture |
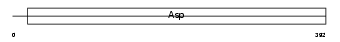
|
|||||
| Description | Cathepsin E precursor (EC 3.4.23.34). | |||||
|
CATD_MOUSE
|
||||||
| θ value | 5.79196e-23 (rank : 7) | NC score | 0.800307 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P18242 | Gene names | Ctsd | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin D precursor (EC 3.4.23.5). | |||||
|
RENI1_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 8) | NC score | 0.787490 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06281, P97911, Q543E5, Q62153, Q62154 | Gene names | Ren1, Ren, Ren-1 | |||
|
Domain Architecture |
|
|||||
| Description | Renin-1 precursor (EC 3.4.23.15) (Angiotensinogenase) (Kidney renin). | |||||
|
RENI_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 9) | NC score | 0.791092 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00797, Q6T5C2 | Gene names | REN | |||
|
Domain Architecture |
|
|||||
| Description | Renin precursor (EC 3.4.23.15) (Angiotensinogenase). | |||||
|
CATD_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 10) | NC score | 0.803328 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07339, Q6IB57 | Gene names | CTSD | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin D precursor (EC 3.4.23.5) [Contains: Cathepsin D light chain; Cathepsin D heavy chain]. | |||||
|
RENI2_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 11) | NC score | 0.786094 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00796, P70229, P97955, Q62155 | Gene names | Ren2, Ren-2 | |||
|
Domain Architecture |
|
|||||
| Description | Renin-2 precursor (EC 3.4.23.15) (Angiotensinogenase) (Submandibular gland renin) [Contains: Renin-2 heavy chain; Renin-2 light chain]. | |||||
|
PEPA_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 12) | NC score | 0.804578 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00790, Q8N1E3 | Gene names | PGA3 | |||
|
Domain Architecture |
|
|||||
| Description | Pepsin A precursor (EC 3.4.23.1). | |||||
|
CATE_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 13) | NC score | 0.801213 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14091, Q9NY58 | Gene names | CTSE | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin E precursor (EC 3.4.23.34). | |||||
|
NAPSA_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 14) | NC score | 0.770008 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O09043 | Gene names | Napsa, Kdap, Nap | |||
|
Domain Architecture |
|
|||||
| Description | Napsin-A precursor (EC 3.4.23.-) (Kidney-derived aspartic protease- like protein) (KDAP-1) (KAP). | |||||
|
NAPSA_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 15) | NC score | 0.754626 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O96009 | Gene names | NAPSA, NAP1 | |||
|
Domain Architecture |
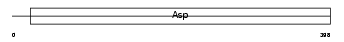
|
|||||
| Description | Napsin-A precursor (EC 3.4.23.-) (Napsin-1) (NAPA) (TA01/TA02) (Aspartyl protease 4) (Asp 4) (ASP4). | |||||
|
WDR6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.028956 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NNW5, Q9UF63 | Gene names | WDR6 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 6. | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.016012 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
K0310_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.021083 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
NDUB8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.016712 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95169, Q5W144, Q9UG53, Q9UJR4, Q9UQF3 | Gene names | NDUFB8 | |||
|
Domain Architecture |
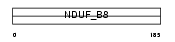
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase ASHI subunit) (Complex I-ASHI) (CI-ASHI). | |||||
|
GDF9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.005715 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07105 | Gene names | Gdf9, Gdf-9 | |||
|
Domain Architecture |

|
|||||
| Description | Growth/differentiation factor 9 precursor (GDF-9). | |||||
|
BACE2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5Z0, Q9UJT6 | Gene names | BACE2, ASP21 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-secretase 2 precursor (EC 3.4.23.45) (Beta-site APP-cleaving enzyme 2) (Aspartyl protease 1) (Asp 1) (ASP1) (Membrane-associated aspartic protease 1) (Memapsin-1) (Down region aspartic protease). | |||||
|
BACE1_MOUSE
|
||||||
| NC score | 0.977177 (rank : 2) | θ value | 3.29472e-127 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56818, Q544D0 | Gene names | Bace1, Bace | |||
|
Domain Architecture |

|
|||||
| Description | Beta-secretase 1 precursor (EC 3.4.23.46) (Beta-site APP cleaving enzyme 1) (Beta-site amyloid precursor protein cleaving enzyme 1) (Membrane-associated aspartic protease 2) (Memapsin-2) (Aspartyl protease 2) (Asp 2) (ASP2). | |||||
|
BACE1_HUMAN
|
||||||
| NC score | 0.976835 (rank : 3) | θ value | 3.29472e-127 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56817, Q9BYB9, Q9BYC0, Q9BYC1, Q9UJT5 | Gene names | BACE1, BACE | |||
|
Domain Architecture |

|
|||||
| Description | Beta-secretase 1 precursor (EC 3.4.23.46) (Beta-site APP cleaving enzyme 1) (Beta-site amyloid precursor protein cleaving enzyme 1) (Membrane-associated aspartic protease 2) (Memapsin-2) (Aspartyl protease 2) (Asp 2) (ASP2). | |||||
|
PEPC_HUMAN
|
||||||
| NC score | 0.810715 (rank : 4) | θ value | 1.12775e-26 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P20142, Q5T3D7, Q5T3D8 | Gene names | PGC | |||
|
Domain Architecture |

|
|||||
| Description | Gastricsin precursor (EC 3.4.23.3) (Pepsinogen C). | |||||
|
PEPA_HUMAN
|
||||||
| NC score | 0.804578 (rank : 5) | θ value | 4.15078e-21 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00790, Q8N1E3 | Gene names | PGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Pepsin A precursor (EC 3.4.23.1). | |||||
|
CATD_HUMAN
|
||||||
| NC score | 0.803328 (rank : 6) | θ value | 3.75424e-22 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07339, Q6IB57 | Gene names | CTSD | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin D precursor (EC 3.4.23.5) [Contains: Cathepsin D light chain; Cathepsin D heavy chain]. | |||||
|
CATE_MOUSE
|
||||||
| NC score | 0.801693 (rank : 7) | θ value | 1.16704e-23 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70269, O35647 | Gene names | Ctse | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin E precursor (EC 3.4.23.34). | |||||
|
PEPC_MOUSE
|
||||||
| NC score | 0.801231 (rank : 8) | θ value | 4.01107e-24 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D7R7, Q9D7T2 | Gene names | Pgc, Upg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gastricsin precursor (EC 3.4.23.3) (Pepsinogen C). | |||||
|
CATE_HUMAN
|
||||||
| NC score | 0.801213 (rank : 9) | θ value | 1.5773e-20 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14091, Q9NY58 | Gene names | CTSE | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin E precursor (EC 3.4.23.34). | |||||
|
CATD_MOUSE
|
||||||
| NC score | 0.800307 (rank : 10) | θ value | 5.79196e-23 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P18242 | Gene names | Ctsd | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin D precursor (EC 3.4.23.5). | |||||
|
RENI_HUMAN
|
||||||
| NC score | 0.791092 (rank : 11) | θ value | 2.87452e-22 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00797, Q6T5C2 | Gene names | REN | |||
|
Domain Architecture |

|
|||||
| Description | Renin precursor (EC 3.4.23.15) (Angiotensinogenase). | |||||
|
RENI1_MOUSE
|
||||||
| NC score | 0.787490 (rank : 12) | θ value | 2.87452e-22 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06281, P97911, Q543E5, Q62153, Q62154 | Gene names | Ren1, Ren, Ren-1 | |||
|
Domain Architecture |

|
|||||
| Description | Renin-1 precursor (EC 3.4.23.15) (Angiotensinogenase) (Kidney renin). | |||||
|
RENI2_MOUSE
|
||||||
| NC score | 0.786094 (rank : 13) | θ value | 1.42661e-21 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00796, P70229, P97955, Q62155 | Gene names | Ren2, Ren-2 | |||
|
Domain Architecture |

|
|||||
| Description | Renin-2 precursor (EC 3.4.23.15) (Angiotensinogenase) (Submandibular gland renin) [Contains: Renin-2 heavy chain; Renin-2 light chain]. | |||||
|
NAPSA_MOUSE
|
||||||
| NC score | 0.770008 (rank : 14) | θ value | 2.13179e-17 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O09043 | Gene names | Napsa, Kdap, Nap | |||
|
Domain Architecture |

|
|||||
| Description | Napsin-A precursor (EC 3.4.23.-) (Kidney-derived aspartic protease- like protein) (KDAP-1) (KAP). | |||||
|
NAPSA_HUMAN
|
||||||
| NC score | 0.754626 (rank : 15) | θ value | 4.91457e-14 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O96009 | Gene names | NAPSA, NAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Napsin-A precursor (EC 3.4.23.-) (Napsin-1) (NAPA) (TA01/TA02) (Aspartyl protease 4) (Asp 4) (ASP4). | |||||
|
WDR6_HUMAN
|
||||||
| NC score | 0.028956 (rank : 16) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NNW5, Q9UF63 | Gene names | WDR6 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 6. | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.021083 (rank : 17) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
NDUB8_HUMAN
|
||||||
| NC score | 0.016712 (rank : 18) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95169, Q5W144, Q9UG53, Q9UJR4, Q9UQF3 | Gene names | NDUFB8 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase ASHI subunit) (Complex I-ASHI) (CI-ASHI). | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.016012 (rank : 19) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
GDF9_MOUSE
|
||||||
| NC score | 0.005715 (rank : 20) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07105 | Gene names | Gdf9, Gdf-9 | |||
|
Domain Architecture |

|
|||||
| Description | Growth/differentiation factor 9 precursor (GDF-9). | |||||