Please be patient as the page loads
|
AT5F1_MOUSE
|
||||||
| SwissProt Accessions | Q9CQQ7 | Gene names | Atp5f1 | |||
|
Domain Architecture |
|
|||||
| Description | ATP synthase B chain, mitochondrial precursor (EC 3.6.3.14). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AT5F1_MOUSE
|
||||||
| θ value | 3.62588e-142 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CQQ7 | Gene names | Atp5f1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP synthase B chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
AT5F1_HUMAN
|
||||||
| θ value | 3.30237e-119 (rank : 2) | NC score | 0.967958 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24539, Q9BQ68, Q9BRU8 | Gene names | ATP5F1 | |||
|
Domain Architecture |
|
|||||
| Description | ATP synthase B chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 3) | NC score | 0.056233 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
FEZ2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 4) | NC score | 0.122519 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6TYB5 | Gene names | Fez2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 2 (Zygin-2) (Zygin II). | |||||
|
FEZ2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 5) | NC score | 0.123398 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UHY8, Q76LN0, Q99690 | Gene names | FEZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 2 (Zygin-2) (Zygin II). | |||||
|
K0555_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 6) | NC score | 0.070795 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
MINK1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.024047 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MINK1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.024177 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
CCD93_MOUSE
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.086133 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TQK5, Q3TX53 | Gene names | Ccdc93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.056230 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
ABCD2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.050164 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBJ2, Q13210 | Gene names | ABCD2, ALD1, ALDL1, ALDR, ALDRP | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family D member 2 (Adrenoleukodystrophy- related protein) (hALDR) (Adrenoleukodystrophy-like 1). | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.062681 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.052454 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
K1C17_MOUSE
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.031931 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QWL7, Q61783 | Gene names | Krt17, Krt1-17 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17). | |||||
|
CTGE4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.046719 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IX94, O95046 | Gene names | CTAGE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 4 (cTAGE-4 protein). | |||||
|
XE7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.054599 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
CJ118_MOUSE
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.063217 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.031560 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.033682 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.036336 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
ABCD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.044271 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33897, Q6GTZ2 | Gene names | ABCD1, ALD | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family D member 1 (Adrenoleukodystrophy protein) (ALDP). | |||||
|
GBP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.036489 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P32456, Q86TB0 | Gene names | GBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (HuGBP-2). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.056055 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.048660 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
K1C17_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.029177 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q04695 | Gene names | KRT17 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17) (39.1). | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.048742 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
TAXB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.047274 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
FOXE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.008605 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00358, O75765 | Gene names | FOXE1, FKHL15, TITF2, TTF2 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein E1 (Thyroid transcription factor 2) (TTF-2) (Forkhead-related protein FKHL15). | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.046703 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
NAB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.025997 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13506, O75383, O75384, Q6GTU1, Q9UEV1 | Gene names | NAB1 | |||
|
Domain Architecture |
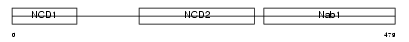
|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1) (Transcriptional regulatory protein p54). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.053251 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
ABCD2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.042214 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61285, Q8BQ63, Q8C4B6 | Gene names | Abcd2, Aldr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family D member 2 (Adrenoleukodystrophy- related protein). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.045921 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.017111 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
GBP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.034697 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P32455 | Gene names | GBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (HuGBP-1). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.035543 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.021025 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.051135 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
CCD91_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.046415 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.047168 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
GNA14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.008696 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30677 | Gene names | Gna14, Gna-14 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein alpha-14 subunit. | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.035623 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.028166 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MOES_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.023491 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.039865 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
FEZ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.059587 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99689, O00679, O00728, Q6IBI7 | Gene names | FEZ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
FEZ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.057743 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K0X8 | Gene names | Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
AT5F1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.62588e-142 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CQQ7 | Gene names | Atp5f1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP synthase B chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
AT5F1_HUMAN
|
||||||
| NC score | 0.967958 (rank : 2) | θ value | 3.30237e-119 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24539, Q9BQ68, Q9BRU8 | Gene names | ATP5F1 | |||
|
Domain Architecture |
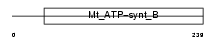
|
|||||
| Description | ATP synthase B chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
FEZ2_HUMAN
|
||||||
| NC score | 0.123398 (rank : 3) | θ value | 0.0330416 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UHY8, Q76LN0, Q99690 | Gene names | FEZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 2 (Zygin-2) (Zygin II). | |||||
|
FEZ2_MOUSE
|
||||||
| NC score | 0.122519 (rank : 4) | θ value | 0.0252991 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6TYB5 | Gene names | Fez2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 2 (Zygin-2) (Zygin II). | |||||
|
CCD93_MOUSE
|
||||||
| NC score | 0.086133 (rank : 5) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TQK5, Q3TX53 | Gene names | Ccdc93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.070795 (rank : 6) | θ value | 0.0736092 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.063217 (rank : 7) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.062681 (rank : 8) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
FEZ1_HUMAN
|
||||||
| NC score | 0.059587 (rank : 9) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99689, O00679, O00728, Q6IBI7 | Gene names | FEZ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
FEZ1_MOUSE
|
||||||
| NC score | 0.057743 (rank : 10) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K0X8 | Gene names | Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.056233 (rank : 11) | θ value | 0.0148317 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.056230 (rank : 12) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.056055 (rank : 13) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.054599 (rank : 14) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.053251 (rank : 15) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.052454 (rank : 16) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.051135 (rank : 17) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
ABCD2_HUMAN
|
||||||
| NC score | 0.050164 (rank : 18) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBJ2, Q13210 | Gene names | ABCD2, ALD1, ALDL1, ALDR, ALDRP | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family D member 2 (Adrenoleukodystrophy- related protein) (hALDR) (Adrenoleukodystrophy-like 1). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.048742 (rank : 19) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.048660 (rank : 20) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
TAXB1_MOUSE
|
||||||
| NC score | 0.047274 (rank : 21) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.047168 (rank : 22) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
CTGE4_HUMAN
|
||||||
| NC score | 0.046719 (rank : 23) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IX94, O95046 | Gene names | CTAGE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 4 (cTAGE-4 protein). | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.046703 (rank : 24) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.046415 (rank : 25) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.045921 (rank : 26) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
ABCD1_HUMAN
|
||||||
| NC score | 0.044271 (rank : 27) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33897, Q6GTZ2 | Gene names | ABCD1, ALD | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family D member 1 (Adrenoleukodystrophy protein) (ALDP). | |||||
|
ABCD2_MOUSE
|
||||||
| NC score | 0.042214 (rank : 28) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61285, Q8BQ63, Q8C4B6 | Gene names | Abcd2, Aldr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family D member 2 (Adrenoleukodystrophy- related protein). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.039865 (rank : 29) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
GBP2_HUMAN
|
||||||
| NC score | 0.036489 (rank : 30) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P32456, Q86TB0 | Gene names | GBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (HuGBP-2). | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.036336 (rank : 31) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.035623 (rank : 32) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.035543 (rank : 33) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
GBP1_HUMAN
|
||||||
| NC score | 0.034697 (rank : 34) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P32455 | Gene names | GBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (HuGBP-1). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.033682 (rank : 35) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
K1C17_MOUSE
|
||||||
| NC score | 0.031931 (rank : 36) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QWL7, Q61783 | Gene names | Krt17, Krt1-17 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.031560 (rank : 37) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
K1C17_HUMAN
|
||||||
| NC score | 0.029177 (rank : 38) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q04695 | Gene names | KRT17 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17) (39.1). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.028166 (rank : 39) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
NAB1_HUMAN
|
||||||
| NC score | 0.025997 (rank : 40) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13506, O75383, O75384, Q6GTU1, Q9UEV1 | Gene names | NAB1 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1) (Transcriptional regulatory protein p54). | |||||
|
MINK1_MOUSE
|
||||||
| NC score | 0.024177 (rank : 41) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MINK1_HUMAN
|
||||||
| NC score | 0.024047 (rank : 42) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.023491 (rank : 43) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.021025 (rank : 44) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.017111 (rank : 45) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
GNA14_MOUSE
|
||||||
| NC score | 0.008696 (rank : 46) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30677 | Gene names | Gna14, Gna-14 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein alpha-14 subunit. | |||||
|
FOXE1_HUMAN
|
||||||
| NC score | 0.008605 (rank : 47) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00358, O75765 | Gene names | FOXE1, FKHL15, TITF2, TTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E1 (Thyroid transcription factor 2) (TTF-2) (Forkhead-related protein FKHL15). | |||||