Please be patient as the page loads
|
APOD_MOUSE
|
||||||
| SwissProt Accessions | P51910 | Gene names | Apod | |||
|
Domain Architecture |
|
|||||
| Description | Apolipoprotein D precursor (Apo-D) (ApoD). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
APOD_MOUSE
|
||||||
| θ value | 9.63072e-111 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51910 | Gene names | Apod | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein D precursor (Apo-D) (ApoD). | |||||
|
APOD_HUMAN
|
||||||
| θ value | 1.65806e-78 (rank : 2) | NC score | 0.989013 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05090 | Gene names | APOD | |||
|
Domain Architecture |
|
|||||
| Description | Apolipoprotein D precursor (Apo-D) (ApoD). | |||||
|
RETBP_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 3) | NC score | 0.383230 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02753, O43478, O43479, Q5VY24, Q8WWA3, Q9P178 | Gene names | RBP4 | |||
|
Domain Architecture |
|
|||||
| Description | Plasma retinol-binding protein precursor (PRBP) (RBP) [Contains: Plasma retinol-binding protein(1-182); Plasma retinol-binding protein(1-181); Plasma retinol-binding protein(1-179); Plasma retinol- binding protein(1-176)]. | |||||
|
RETBP_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 4) | NC score | 0.379516 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00724, P70357, Q566I5 | Gene names | Rbp4 | |||
|
Domain Architecture |
|
|||||
| Description | Plasma retinol-binding protein precursor (PRBP) (RBP). | |||||
|
PTGDS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 5) | NC score | 0.102627 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41222, Q5SQ10, Q7M4P3, Q9UCC9, Q9UCD9 | Gene names | PTGDS, PDS | |||
|
Domain Architecture |
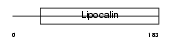
|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS) (Beta-trace protein) (Cerebrin-28). | |||||
|
LCN6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 6) | NC score | 0.074612 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62502, Q71SF6 | Gene names | LCN6, LCN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-6 precursor (Lipocalin-5). | |||||
|
NGAL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 7) | NC score | 0.058421 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11672 | Gene names | Lcn2 | |||
|
Domain Architecture |
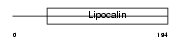
|
|||||
| Description | Neutrophil gelatinase-associated lipocalin precursor (NGAL) (p25) (SV- 40-induced 24P3 protein) (Lipocalin-2). | |||||
|
AMBP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.056065 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |
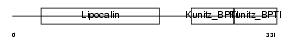
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
LCN12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.057617 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6JVE5 | Gene names | LCN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-12 precursor. | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 10) | NC score | 0.021801 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
K0776_HUMAN
|
||||||
| θ value | 8.99809 (rank : 11) | NC score | 0.029349 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94874, Q8N765, Q9NTQ0 | Gene names | KIAA0776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0776. | |||||
|
K0776_MOUSE
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.029248 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CCJ3, Q3V145, Q6ZQ50, Q8BT70, Q8C484, Q9D8I8 | Gene names | Kiaa0776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0776. | |||||
|
APOD_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 9.63072e-111 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51910 | Gene names | Apod | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein D precursor (Apo-D) (ApoD). | |||||
|
APOD_HUMAN
|
||||||
| NC score | 0.989013 (rank : 2) | θ value | 1.65806e-78 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05090 | Gene names | APOD | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein D precursor (Apo-D) (ApoD). | |||||
|
RETBP_HUMAN
|
||||||
| NC score | 0.383230 (rank : 3) | θ value | 0.000158464 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02753, O43478, O43479, Q5VY24, Q8WWA3, Q9P178 | Gene names | RBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma retinol-binding protein precursor (PRBP) (RBP) [Contains: Plasma retinol-binding protein(1-182); Plasma retinol-binding protein(1-181); Plasma retinol-binding protein(1-179); Plasma retinol- binding protein(1-176)]. | |||||
|
RETBP_MOUSE
|
||||||
| NC score | 0.379516 (rank : 4) | θ value | 0.000270298 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00724, P70357, Q566I5 | Gene names | Rbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma retinol-binding protein precursor (PRBP) (RBP). | |||||
|
PTGDS_HUMAN
|
||||||
| NC score | 0.102627 (rank : 5) | θ value | 1.81305 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41222, Q5SQ10, Q7M4P3, Q9UCC9, Q9UCD9 | Gene names | PTGDS, PDS | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS) (Beta-trace protein) (Cerebrin-28). | |||||
|
LCN6_HUMAN
|
||||||
| NC score | 0.074612 (rank : 6) | θ value | 2.36792 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62502, Q71SF6 | Gene names | LCN6, LCN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-6 precursor (Lipocalin-5). | |||||
|
NGAL_MOUSE
|
||||||
| NC score | 0.058421 (rank : 7) | θ value | 2.36792 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11672 | Gene names | Lcn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil gelatinase-associated lipocalin precursor (NGAL) (p25) (SV- 40-induced 24P3 protein) (Lipocalin-2). | |||||
|
LCN12_HUMAN
|
||||||
| NC score | 0.057617 (rank : 8) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6JVE5 | Gene names | LCN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-12 precursor. | |||||
|
AMBP_HUMAN
|
||||||
| NC score | 0.056065 (rank : 9) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
K0776_HUMAN
|
||||||
| NC score | 0.029349 (rank : 10) | θ value | 8.99809 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94874, Q8N765, Q9NTQ0 | Gene names | KIAA0776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0776. | |||||
|
K0776_MOUSE
|
||||||
| NC score | 0.029248 (rank : 11) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CCJ3, Q3V145, Q6ZQ50, Q8BT70, Q8C484, Q9D8I8 | Gene names | Kiaa0776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0776. | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.021801 (rank : 12) | θ value | 5.27518 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||