Please be patient as the page loads
|
AL1A2_MOUSE
|
||||||
| SwissProt Accessions | Q62148, Q6DI79 | Gene names | Aldh1a2, Aldh1a7, Raldh2 | |||
|
Domain Architecture |
|
|||||
| Description | Retinal dehydrogenase 2 (EC 1.2.1.36) (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AL1A1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998651 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P00352, O00768 | Gene names | ALDH1A1, ALDC, ALDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 1 (EC 1.2.1.36) (RalDH1) (RALDH 1) (Aldehyde dehydrogenase family 1 member A1) (Aldehyde dehydrogenase, cytosolic) (ALHDII) (ALDH-E1). | |||||
|
AL1A1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998715 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P24549, Q7TQJ0, Q811J0 | Gene names | Aldh1a1, Ahd-2, Ahd2, Aldh1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 1 (EC 1.2.1.36) (RalDH1) (RALDH 1) (Aldehyde dehydrogenase family 1 member A1) (Aldehyde dehydrogenase, cytosolic) (ALHDII) (ALDH-E1). | |||||
|
AL1A2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999886 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O94788, Q2PJS6, Q8NHQ4, Q9UBR8, Q9UFY0 | Gene names | ALDH1A2, RALDH2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 2 (EC 1.2.1.36) (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2). | |||||
|
AL1A2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62148, Q6DI79 | Gene names | Aldh1a2, Aldh1a7, Raldh2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 2 (EC 1.2.1.36) (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2). | |||||
|
AL1A3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.998251 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47895, Q6NT64 | Gene names | ALDH1A3, ALDH6 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase 1A3 (EC 1.2.1.5) (Aldehyde dehydrogenase 6) (Retinaldehyde dehydrogenase 3) (RALDH-3). | |||||
|
AL1A3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.998089 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JHW9, Q9EQP7, Q9JI72 | Gene names | Aldh1a3, Aldh6, Raldh3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aldehyde dehydrogenase 1A3 (EC 1.2.1.5) (Aldehyde dehydrogenase 6) (Retinaldehyde dehydrogenase 3) (RALDH-3). | |||||
|
AL1B1_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.998516 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30837 | Gene names | ALDH1B1, ALDH5, ALDHX | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase X, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2). | |||||
|
ALDH2_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.998110 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05091, Q03639, Q6IB13, Q6IV71 | Gene names | ALDH2, ALDM | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2) (ALDHI) (ALDH-E2). | |||||
|
ALDH2_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.998168 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47738 | Gene names | Aldh2, Ahd-1, Ahd1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2) (AHD-M1) (ALDHI) (ALDH-E2). | |||||
|
FTHFD_HUMAN
|
||||||
| θ value | 7.57797e-132 (rank : 10) | NC score | 0.976061 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75891, Q68CS1 | Gene names | ALDH1L1, FTHFD | |||
|
Domain Architecture |

|
|||||
| Description | 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) (10-FTHFDH) (Aldehyde dehydrogenase 1 family member L1). | |||||
|
FTHFD_MOUSE
|
||||||
| θ value | 4.9119e-131 (rank : 11) | NC score | 0.974463 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R0Y6 | Gene names | Aldh1l1, Fthfd | |||
|
Domain Architecture |

|
|||||
| Description | 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) (10-FTHFDH) (Aldehyde dehydrogenase 1 family member L1). | |||||
|
AL9A1_HUMAN
|
||||||
| θ value | 1.26366e-94 (rank : 12) | NC score | 0.976917 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49189, Q5VV90, Q6LCL1, Q9NZT7 | Gene names | ALDH9A1, ALDH7, ALDH9 | |||
|
Domain Architecture |

|
|||||
| Description | 4-trimethylaminobutyraldehyde dehydrogenase (EC 1.2.1.47) (TMABADH) (Aldehyde dehydrogenase 9A1) (EC 1.2.1.3) (Aldehyde dehydrogenase E3 isozyme) (Gamma-aminobutyraldehyde dehydrogenase) (EC 1.2.1.19) (R- aminobutyraldehyde dehydrogenase). | |||||
|
AL9A1_MOUSE
|
||||||
| θ value | 3.80478e-91 (rank : 13) | NC score | 0.976118 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JLJ2 | Gene names | Aldh9a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 4-trimethylaminobutyraldehyde dehydrogenase (EC 1.2.1.47) (TMABADH) (Aldehyde dehydrogenase 9A1) (EC 1.2.1.3). | |||||
|
SSDH_HUMAN
|
||||||
| θ value | 2.47765e-74 (rank : 14) | NC score | 0.964468 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51649 | Gene names | ALDH5A1, SSADH | |||
|
Domain Architecture |

|
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
SSDH_MOUSE
|
||||||
| θ value | 5.51964e-74 (rank : 15) | NC score | 0.964591 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BWF0 | Gene names | Aldh5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
MMSA_HUMAN
|
||||||
| θ value | 3.36421e-55 (rank : 16) | NC score | 0.963875 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02252, Q9UKM8 | Gene names | ALDH6A1, MMSDH | |||
|
Domain Architecture |

|
|||||
| Description | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial precursor (EC 1.2.1.27) (MMSDH) (Malonate-semialdehyde dehydrogenase [acylating]) (EC 1.2.1.18). | |||||
|
AL7A1_HUMAN
|
||||||
| θ value | 8.28633e-54 (rank : 17) | NC score | 0.942516 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49419, O14619, Q6IPU8, Q9BUL4 | Gene names | ALDH7A1, ATQ1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase family 7 member A1 (EC 1.2.1.3) (Antiquitin-1). | |||||
|
AL7A1_MOUSE
|
||||||
| θ value | 7.25919e-50 (rank : 18) | NC score | 0.938251 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DBF1 | Gene names | Aldh7a1, Ald7a1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase family 7 member A1 (EC 1.2.1.3) (Antiquitin-1). | |||||
|
AL3A1_HUMAN
|
||||||
| θ value | 1.73584e-35 (rank : 19) | NC score | 0.845770 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30838, Q9BT37 | Gene names | ALDH3A1, ALDH3 | |||
|
Domain Architecture |
|
|||||
| Description | Aldehyde dehydrogenase, dimeric NADP-preferring (EC 1.2.1.5) (ALDH class 3) (ALDHIII). | |||||
|
AL4A1_HUMAN
|
||||||
| θ value | 2.50655e-34 (rank : 20) | NC score | 0.888892 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30038, Q16882, Q8IZ38, Q96IF0 | Gene names | ALDH4A1, ALDH4, P5CDH | |||
|
Domain Architecture |

|
|||||
| Description | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial precursor (EC 1.5.1.12) (P5C dehydrogenase) (Aldehyde dehydrogenase 4A1). | |||||
|
AL3A1_MOUSE
|
||||||
| θ value | 5.584e-34 (rank : 21) | NC score | 0.844857 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47739, Q9R203 | Gene names | Aldh3a1, Ahd-4, Ahd4, Aldh3, Aldh4 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, dimeric NADP-preferring (EC 1.2.1.5) (ALDH class 3) (Dioxin-inducible aldehyde dehydrogenase 3). | |||||
|
AL4A1_MOUSE
|
||||||
| θ value | 2.12192e-33 (rank : 22) | NC score | 0.880489 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CHT0, Q7TND0, Q8BXM3, Q8R0N1, Q8R1S2 | Gene names | Aldh4a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial precursor (EC 1.5.1.12) (P5C dehydrogenase) (Aldehyde dehydrogenase 4A1). | |||||
|
AL3A2_MOUSE
|
||||||
| θ value | 6.82597e-32 (rank : 23) | NC score | 0.839591 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47740 | Gene names | Aldh3a2, Ahd-3, Ahd3, Aldh3, Aldh4 | |||
|
Domain Architecture |
|
|||||
| Description | Fatty aldehyde dehydrogenase (EC 1.2.1.3) (Aldehyde dehydrogenase, microsomal) (Aldehyde dehydrogenase family 3 member A2) (Aldehyde dehydrogenase 10). | |||||
|
AL3A2_HUMAN
|
||||||
| θ value | 7.54701e-31 (rank : 24) | NC score | 0.828076 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51648, Q93011, Q96J37 | Gene names | ALDH3A2, ALDH10, FALDH | |||
|
Domain Architecture |

|
|||||
| Description | Fatty aldehyde dehydrogenase (EC 1.2.1.3) (Aldehyde dehydrogenase, microsomal) (Aldehyde dehydrogenase family 3 member A2) (Aldehyde dehydrogenase 10). | |||||
|
AL3B1_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 25) | NC score | 0.821808 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43353, Q53XL5, Q8N515, Q96CK8 | Gene names | ALDH3B1, ALDH7 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase 3B1 (EC 1.2.1.5) (Aldehyde dehydrogenase 7). | |||||
|
AL3B1_MOUSE
|
||||||
| θ value | 1.2049e-28 (rank : 26) | NC score | 0.822223 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80VQ0, Q63ZW3, Q8VHW0, Q9CW05 | Gene names | Aldh3b1, Aldh7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aldehyde dehydrogenase 3B1 (EC 1.2.1.5) (Aldehyde dehydrogenase 7). | |||||
|
AL3B2_HUMAN
|
||||||
| θ value | 2.05525e-28 (rank : 27) | NC score | 0.834583 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48448, Q8NAL5, Q96IB2 | Gene names | ALDH3B2, ALDH8 | |||
|
Domain Architecture |
|
|||||
| Description | Aldehyde dehydrogenase 3B2 (EC 1.2.1.5) (Aldehyde dehydrogenase 8). | |||||
|
MAOX_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.017632 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06801 | Gene names | Me1, Mod-1, Mod1 | |||
|
Domain Architecture |

|
|||||
| Description | NADP-dependent malic enzyme (EC 1.1.1.40) (NADP-ME) (Malic enzyme 1). | |||||
|
CD4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.015737 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06332 | Gene names | Cd4 | |||
|
Domain Architecture |
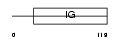
|
|||||
| Description | T-cell surface glycoprotein CD4 precursor (T-cell surface antigen T4/Leu-3) (T-cell differentiation antigen L3T4). | |||||
|
MKS3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.007053 (rank : 33) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BR76, Q78U07 | Gene names | Tmem67, Mks3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Meckelin (Meckel syndrome type 3 protein homolog) (Transmembrane protein 67). | |||||
|
MAOX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.009621 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48163, Q16797, Q16855, Q53F72, Q5VWA2, Q9BWX8, Q9H1W3, Q9UIY4 | Gene names | ME1 | |||
|
Domain Architecture |

|
|||||
| Description | NADP-dependent malic enzyme (EC 1.1.1.40) (NADP-ME) (Malic enzyme 1). | |||||
|
TLR12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.000751 (rank : 34) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6QNU9 | Gene names | Tlr12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Toll-like receptor 12 precursor. | |||||
|
FMT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.074047 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96DP5 | Gene names | MTFMT, FMT, FMT1 | |||
|
Domain Architecture |
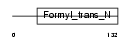
|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
FMT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.077159 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D799, Q8VE89 | Gene names | Mtfmt, Fmt | |||
|
Domain Architecture |
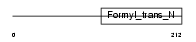
|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
AL1A2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62148, Q6DI79 | Gene names | Aldh1a2, Aldh1a7, Raldh2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 2 (EC 1.2.1.36) (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2). | |||||
|
AL1A2_HUMAN
|
||||||
| NC score | 0.999886 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O94788, Q2PJS6, Q8NHQ4, Q9UBR8, Q9UFY0 | Gene names | ALDH1A2, RALDH2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 2 (EC 1.2.1.36) (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2). | |||||
|
AL1A1_MOUSE
|
||||||
| NC score | 0.998715 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P24549, Q7TQJ0, Q811J0 | Gene names | Aldh1a1, Ahd-2, Ahd2, Aldh1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 1 (EC 1.2.1.36) (RalDH1) (RALDH 1) (Aldehyde dehydrogenase family 1 member A1) (Aldehyde dehydrogenase, cytosolic) (ALHDII) (ALDH-E1). | |||||
|
AL1A1_HUMAN
|
||||||
| NC score | 0.998651 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P00352, O00768 | Gene names | ALDH1A1, ALDC, ALDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 1 (EC 1.2.1.36) (RalDH1) (RALDH 1) (Aldehyde dehydrogenase family 1 member A1) (Aldehyde dehydrogenase, cytosolic) (ALHDII) (ALDH-E1). | |||||
|
AL1B1_HUMAN
|
||||||
| NC score | 0.998516 (rank : 5) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30837 | Gene names | ALDH1B1, ALDH5, ALDHX | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase X, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2). | |||||
|
AL1A3_HUMAN
|
||||||
| NC score | 0.998251 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47895, Q6NT64 | Gene names | ALDH1A3, ALDH6 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase 1A3 (EC 1.2.1.5) (Aldehyde dehydrogenase 6) (Retinaldehyde dehydrogenase 3) (RALDH-3). | |||||
|
ALDH2_MOUSE
|
||||||
| NC score | 0.998168 (rank : 7) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47738 | Gene names | Aldh2, Ahd-1, Ahd1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2) (AHD-M1) (ALDHI) (ALDH-E2). | |||||
|
ALDH2_HUMAN
|
||||||
| NC score | 0.998110 (rank : 8) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05091, Q03639, Q6IB13, Q6IV71 | Gene names | ALDH2, ALDM | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2) (ALDHI) (ALDH-E2). | |||||
|
AL1A3_MOUSE
|
||||||
| NC score | 0.998089 (rank : 9) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JHW9, Q9EQP7, Q9JI72 | Gene names | Aldh1a3, Aldh6, Raldh3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aldehyde dehydrogenase 1A3 (EC 1.2.1.5) (Aldehyde dehydrogenase 6) (Retinaldehyde dehydrogenase 3) (RALDH-3). | |||||
|
AL9A1_HUMAN
|
||||||
| NC score | 0.976917 (rank : 10) | θ value | 1.26366e-94 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49189, Q5VV90, Q6LCL1, Q9NZT7 | Gene names | ALDH9A1, ALDH7, ALDH9 | |||
|
Domain Architecture |

|
|||||
| Description | 4-trimethylaminobutyraldehyde dehydrogenase (EC 1.2.1.47) (TMABADH) (Aldehyde dehydrogenase 9A1) (EC 1.2.1.3) (Aldehyde dehydrogenase E3 isozyme) (Gamma-aminobutyraldehyde dehydrogenase) (EC 1.2.1.19) (R- aminobutyraldehyde dehydrogenase). | |||||
|
AL9A1_MOUSE
|
||||||
| NC score | 0.976118 (rank : 11) | θ value | 3.80478e-91 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JLJ2 | Gene names | Aldh9a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 4-trimethylaminobutyraldehyde dehydrogenase (EC 1.2.1.47) (TMABADH) (Aldehyde dehydrogenase 9A1) (EC 1.2.1.3). | |||||
|
FTHFD_HUMAN
|
||||||
| NC score | 0.976061 (rank : 12) | θ value | 7.57797e-132 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75891, Q68CS1 | Gene names | ALDH1L1, FTHFD | |||
|
Domain Architecture |

|
|||||
| Description | 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) (10-FTHFDH) (Aldehyde dehydrogenase 1 family member L1). | |||||
|
FTHFD_MOUSE
|
||||||
| NC score | 0.974463 (rank : 13) | θ value | 4.9119e-131 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R0Y6 | Gene names | Aldh1l1, Fthfd | |||
|
Domain Architecture |

|
|||||
| Description | 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) (10-FTHFDH) (Aldehyde dehydrogenase 1 family member L1). | |||||
|
SSDH_MOUSE
|
||||||
| NC score | 0.964591 (rank : 14) | θ value | 5.51964e-74 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BWF0 | Gene names | Aldh5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
SSDH_HUMAN
|
||||||
| NC score | 0.964468 (rank : 15) | θ value | 2.47765e-74 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51649 | Gene names | ALDH5A1, SSADH | |||
|
Domain Architecture |

|
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
MMSA_HUMAN
|
||||||
| NC score | 0.963875 (rank : 16) | θ value | 3.36421e-55 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02252, Q9UKM8 | Gene names | ALDH6A1, MMSDH | |||
|
Domain Architecture |

|
|||||
| Description | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial precursor (EC 1.2.1.27) (MMSDH) (Malonate-semialdehyde dehydrogenase [acylating]) (EC 1.2.1.18). | |||||
|
AL7A1_HUMAN
|
||||||
| NC score | 0.942516 (rank : 17) | θ value | 8.28633e-54 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49419, O14619, Q6IPU8, Q9BUL4 | Gene names | ALDH7A1, ATQ1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase family 7 member A1 (EC 1.2.1.3) (Antiquitin-1). | |||||
|
AL7A1_MOUSE
|
||||||
| NC score | 0.938251 (rank : 18) | θ value | 7.25919e-50 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DBF1 | Gene names | Aldh7a1, Ald7a1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase family 7 member A1 (EC 1.2.1.3) (Antiquitin-1). | |||||
|
AL4A1_HUMAN
|
||||||
| NC score | 0.888892 (rank : 19) | θ value | 2.50655e-34 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30038, Q16882, Q8IZ38, Q96IF0 | Gene names | ALDH4A1, ALDH4, P5CDH | |||
|
Domain Architecture |

|
|||||
| Description | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial precursor (EC 1.5.1.12) (P5C dehydrogenase) (Aldehyde dehydrogenase 4A1). | |||||
|
AL4A1_MOUSE
|
||||||
| NC score | 0.880489 (rank : 20) | θ value | 2.12192e-33 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CHT0, Q7TND0, Q8BXM3, Q8R0N1, Q8R1S2 | Gene names | Aldh4a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial precursor (EC 1.5.1.12) (P5C dehydrogenase) (Aldehyde dehydrogenase 4A1). | |||||
|
AL3A1_HUMAN
|
||||||
| NC score | 0.845770 (rank : 21) | θ value | 1.73584e-35 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30838, Q9BT37 | Gene names | ALDH3A1, ALDH3 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, dimeric NADP-preferring (EC 1.2.1.5) (ALDH class 3) (ALDHIII). | |||||
|
AL3A1_MOUSE
|
||||||
| NC score | 0.844857 (rank : 22) | θ value | 5.584e-34 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47739, Q9R203 | Gene names | Aldh3a1, Ahd-4, Ahd4, Aldh3, Aldh4 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, dimeric NADP-preferring (EC 1.2.1.5) (ALDH class 3) (Dioxin-inducible aldehyde dehydrogenase 3). | |||||
|
AL3A2_MOUSE
|
||||||
| NC score | 0.839591 (rank : 23) | θ value | 6.82597e-32 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47740 | Gene names | Aldh3a2, Ahd-3, Ahd3, Aldh3, Aldh4 | |||
|
Domain Architecture |

|
|||||
| Description | Fatty aldehyde dehydrogenase (EC 1.2.1.3) (Aldehyde dehydrogenase, microsomal) (Aldehyde dehydrogenase family 3 member A2) (Aldehyde dehydrogenase 10). | |||||
|
AL3B2_HUMAN
|
||||||
| NC score | 0.834583 (rank : 24) | θ value | 2.05525e-28 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48448, Q8NAL5, Q96IB2 | Gene names | ALDH3B2, ALDH8 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase 3B2 (EC 1.2.1.5) (Aldehyde dehydrogenase 8). | |||||
|
AL3A2_HUMAN
|
||||||
| NC score | 0.828076 (rank : 25) | θ value | 7.54701e-31 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51648, Q93011, Q96J37 | Gene names | ALDH3A2, ALDH10, FALDH | |||
|
Domain Architecture |

|
|||||
| Description | Fatty aldehyde dehydrogenase (EC 1.2.1.3) (Aldehyde dehydrogenase, microsomal) (Aldehyde dehydrogenase family 3 member A2) (Aldehyde dehydrogenase 10). | |||||
|
AL3B1_MOUSE
|
||||||
| NC score | 0.822223 (rank : 26) | θ value | 1.2049e-28 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80VQ0, Q63ZW3, Q8VHW0, Q9CW05 | Gene names | Aldh3b1, Aldh7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aldehyde dehydrogenase 3B1 (EC 1.2.1.5) (Aldehyde dehydrogenase 7). | |||||
|
AL3B1_HUMAN
|
||||||
| NC score | 0.821808 (rank : 27) | θ value | 1.85889e-29 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43353, Q53XL5, Q8N515, Q96CK8 | Gene names | ALDH3B1, ALDH7 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase 3B1 (EC 1.2.1.5) (Aldehyde dehydrogenase 7). | |||||
|
FMT_MOUSE
|
||||||
| NC score | 0.077159 (rank : 28) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D799, Q8VE89 | Gene names | Mtfmt, Fmt | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
FMT_HUMAN
|
||||||
| NC score | 0.074047 (rank : 29) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96DP5 | Gene names | MTFMT, FMT, FMT1 | |||
|
Domain Architecture |
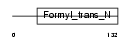
|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
MAOX_MOUSE
|
||||||
| NC score | 0.017632 (rank : 30) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06801 | Gene names | Me1, Mod-1, Mod1 | |||
|
Domain Architecture |

|
|||||
| Description | NADP-dependent malic enzyme (EC 1.1.1.40) (NADP-ME) (Malic enzyme 1). | |||||
|
CD4_MOUSE
|
||||||
| NC score | 0.015737 (rank : 31) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06332 | Gene names | Cd4 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD4 precursor (T-cell surface antigen T4/Leu-3) (T-cell differentiation antigen L3T4). | |||||
|
MAOX_HUMAN
|
||||||
| NC score | 0.009621 (rank : 32) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48163, Q16797, Q16855, Q53F72, Q5VWA2, Q9BWX8, Q9H1W3, Q9UIY4 | Gene names | ME1 | |||
|
Domain Architecture |

|
|||||
| Description | NADP-dependent malic enzyme (EC 1.1.1.40) (NADP-ME) (Malic enzyme 1). | |||||
|
MKS3_MOUSE
|
||||||
| NC score | 0.007053 (rank : 33) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BR76, Q78U07 | Gene names | Tmem67, Mks3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Meckelin (Meckel syndrome type 3 protein homolog) (Transmembrane protein 67). | |||||
|
TLR12_MOUSE
|
||||||
| NC score | 0.000751 (rank : 34) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6QNU9 | Gene names | Tlr12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Toll-like receptor 12 precursor. | |||||